Abstract
TANK-binding kinase 1 (TBK1) responds to microbial stimuli and mediates the induction of type I interferon (IFN). Here, we show that TBK1 is also a central mediator of growth factor signalling; this function of TBK1 relies on a specific adaptor—TBK-binding protein 1 (TBKBP1). TBKBP1 recruits TBK1 to protein kinase C-theta (PKCθ) through a scaffold protein, CARD10. This enables PKCθ to phosphorylate TBK1 at Ser 716, a crucial step for TBK1 activation by growth factors but not by innate immune stimuli. Although the TBK1–TBKBP1 signalling axis is not required for the induction of type I IFN, it mediates mTORC1 activation and oncogenesis. Conditional deletion of either TBK1 or TBKBP1 in lung epithelial cells inhibits tumourigenesis in a mouse model of lung cancer. In addition to promoting tumour growth, the TBK1–TBKBP1 axis facilitates tumour-mediated immunosuppression through a mechanism that involves induction of the checkpoint molecule PD-L1 and stimulation of glycolysis. These findings suggest a PKCθ–TBKBP1–TBK1 growth factor signalling axis that mediates both tumour growth and immunosuppression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Source data are available online for Figs. 1e–g,i,k, 3i,j, 4k,l, 6c–e,g and 7a,c,e–g,i–k, and Extended Data Figs. 1b–e and 7a–c,g, and unprocessed blots are provided for Figs. 1–7 and Extended Data Figs. 1–6. The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) through the MASSIVE repository (MSV000084505) under the accession code PXD016025. All other data that support the findings of this study are available from the corresponding author on reasonable request.
References
Hiscott, J. Convergence of the NF-κB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18, 483–490 (2007).
Shi, J. H., Xie, X. & Sun, S. C. TBK1 as a regulator of autoimmunity and antitumor immunity. Cell. Mol. Immunol. 15, 743–745 (2018).
Chien, Y. et al. RalB GTPase-mediated activation of the IκB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127, 157–170 (2006).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Ou, Y. H. et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol. Cell 41, 458–470 (2011).
Xie, X. et al. IκB kinase ε and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc. Natl Acad. Sci. USA 108, 6474–6479 (2011).
Cooper, J. M. et al. TBK1 provides context-selective support of the activated AKT/mTOR pathway in lung cancer. Cancer Res. 77, 5077–5094 (2017).
Bodur, C. et al. The IKK-related kinase TBK1 activates mTORC1 directly in response to growth factors and innate immune agonists. EMBO J. 37, 19–38 (2018).
Yu, J. et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat. Commun. 6, 6074 (2015).
Häcker, H. et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207 (2006).
Oganesyan, G. et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439, 208–211 (2006).
Liu, S. et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2, e00785 (2013).
Fang, R. et al. MAVS activates TBK1 and IKKε through TRAFs in NEMO dependent and independent manner. PLoS Pathog. 13, e1006720 (2017).
Ryzhakov, G. & Randow, F. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 26, 3180–3190 (2007).
Goncalves, A. et al. Functional dissection of the TBK1 molecular network. PLoS ONE 6, e23971 (2011).
Kawagoe, T. et al. TANK is a negative regulator of Toll-like receptor signaling and is critical for the prevention of autoimmune nephritis. Nat. Immunol. 10, 965–972 (2009).
Fukasaka, M. et al. Critical role of AZI2 in GM-CSF-induced dendritic cell differentiation. J. Immunol. 190, 5702–5711 (2013).
Cross, M. & Dexter, T. M. Growth factors in development, transformation, and tumorigenesis. Cell 64, 271–280 (1991).
Halper, J. Growth factors as active participants in carcinogenesis: a perspective. Vet. Pathol. 47, 77–97 (2010).
Ardito, C. M. et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell 22, 304–317 (2012).
Johnson, L. et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001).
Joung, S. M. et al. Akt contributes to activation of the TRIF-dependent signaling pathways of TLRs by interacting with TANK-binding kinase 1. J. Immunol. 186, 499–507 (2011).
Roux, K. J., Kim, D. I., Burke, B. & May, D. G. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 91, 19.23.1–19.23.15 (2018).
Oliva, J. L., Griner, E. M. & Kazanietz, M. G. PKC isozymes and diacylglycerol-regulated proteins as effectors of growth factor receptors. Growth Factors 23, 245–252 (2005).
Yang, W. et al. EGFR-induced and PKCε monoubiquitylation-dependent NF-κB activation upregulates PKM2 expression and promotes tumorigenesis. Mol. Cell 48, 771–784 (2012).
Ma, X. et al. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl Acad. Sci. USA 109, 9378–9383 (2012).
Shu, C. et al. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure 21, 1137–1148 (2013).
Hornbeck, P. V., Chabra, I., Kornhauser, J. M., Skrzypek, E. & Zhang, B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 (2004).
Pan, D. et al. MALT1 is required for EGFR-induced NF-κB activation and contributes to EGFR-driven lung cancer progression. Oncogene 35, 919–928 (2016).
Reilly, S. M. et al. An inhibitor of the protein kinases TBK1 and IKK-ε improves obesity-related metabolic dysfunctions in mice. Nat. Med. 19, 313–321 (2013).
DuPage, M. et al. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell 19, 72–85 (2011).
Akbay, E. A. et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 3, 1355–1363 (2013).
Lastwika, K. J. et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 76, 227–238 (2016).
Shishibori, T. et al. Three distinct anti-allergic drugs, amlexanox, cromolyn and tranilast, bind to S100A12 and S100A13 of the S100 protein family. Biochem. J. 338, 583–589 (1999).
Bresnick, A. R., Weber, D. J. & Zimmer, D. B. S100 proteins in cancer. Nat. Rev. Cancer 15, 96–109 (2015).
Cho, C. C., Chou, R. H. & Yu, C. Amlexanox blocks the interaction between S100A4 and epidermal growth factor and inhibits cell proliferation. PLoS ONE 11, e0161663 (2016).
Tamai, H. et al. Amlexanox downregulates S100A6 to sensitize KMT2A/AFF1-positive acute lymphoblastic leukemia to TNFα treatment. Cancer Res. 77, 4426–4433 (2017).
Concha-Benavente, F. & Ferris, R. L. Oncogenic growth factor signaling mediating tumor escape from cellular immunity. Curr. Opin. Immunol. 45, 52–59 (2017).
Murillo, M. M. et al. RAS interaction with PI3K p110α is required for tumor-induced angiogenesis. J. Clin. Invest. 124, 3601–3611 (2014).
Chen, N. et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J. Thorac. Oncol. 10, 910–923 (2015).
Li, W. et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor Immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metab. 28, 87–103 (2018).
Cascone, T. et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 27, 977–987 (2018).
Chang, C. H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015).
Pearce, E. L., Poffenberger, M. C., Chang, C. H. & Jones, R. G. Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013).
Galluzzi, L., Chan, T. A., Kroemer, G., Wolchok, J. D. & Lopez-Soto, A. The hallmarks of successful anticancer immunotherapy. Sci. Transl. Med. 10, eaat7807 (2018).
Parsa, A. T. et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 13, 84–88 (2007).
Marzec, M. et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc. Natl Acad. Sci. USA 105, 20852–20857 (2008).
Xu, C. et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 25, 590–604 (2014).
Wang, B. et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature 545, 365–369 (2017).
Zhang, W. et al. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco. Targets Ther. 10, 763–771 (2017).
Jin, J. et al. The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-κB signaling. Nat. Immunol. 13, 1101–1109 (2012).
Li, H. et al. Cre-mediated recombination in mouse Clara cells. Genesis 46, 300–307 (2008).
Zhu, L. et al. TBK-binding protein 1 regulates IL-15-induced autophagy and NKT cell survival. Nat. Commun. 9, 2812 (2018).
Reiley, W., Zhang, M., Wu, X., Graner, E. & Sun, S.-C. Regulation of the deubiquitinating enzyme CYLD by IκB kinase gamma-dependent phosphorylation. Mol. Cell. Biol. 25, 3886–3895 (2005).
Li, Y. et al. Preventing abnormal NF-κB activation and autoimmunity by Otub1-mediated p100 stabilization. Cell Res. 29, 474–485 (2019).
Chang, M., Jin, W. & Sun, S. C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 10, 1089–1095 (2009).
Reiley, W. W. et al. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat. Immunol. 7, 411–417 (2006).
Acknowledgements
We thank F. Zhu, C. Wang and X. Lin for plasmid DNAs; S. Akira for Tank knockout mice and F. J. DeMayo for the Ccspcre mice; the personnel from the flow cytometry, DNA analysis and animal facilities at The MD Anderson Cancer Center and the Mass Spectrometry Proteomics Core at Baylor College of Medicine for technical assistance. This study was supported by a National Institutes of Health grant (AI057555) and partially supported by a seed fund from the Center for Inflammation and Cancer at the MD Anderson Cancer Center. T.G. was a visiting student supported by a scholarship from the China Scholarship Council (grant number 201906380080). The MD Anderson core facilities are supported by a NIH/NCI Cancer Center Support Grant (CCSG; P30CA016672), and the Baylor College of Medicine Mass Spectrometry Proteomics Core is supported by a CPRIT Core Facility Award (RP170005) and a P30 Cancer Center Support Grant (NCI-CA125123).
Author information
Authors and Affiliations
Contributions
L.Z. designed and performed the research, prepared the figures and wrote part of the manuscript. Y.L., X.X., X.Z., M.G., Z.J., C.-J.K., T.G., B.E.H. and X.C. contributed to the experiments. S.-C.S. supervised the work and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
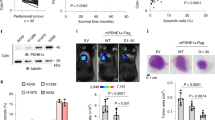
Extended Data Fig. 1 TBKBP1 is not required for TBK1 activation or type I IFN induction by TLR ligands and viruses.
a, Immunoblot analysis of the indicated phosphorylated (P-) and total proteins in whole-cell lysates of A549 cells stably infected with a control shRNA or a Tbkbp1-specific shRNA, stimulated with the indicated inducers. b-e, qRT-PCR analysis of Ifna and Ifnb mRNA expression in A549 cells stably infected with a control shRNA (shCtrl) or shRNAs for Tbk1 (b,c) or Tbkbp1 (d,e), stimulated with the indicated inducers. Data are representative of three independent experiments. n = 3 (b,d,e) or 5 (c) per group. Two-sided unpaired Student’s t-test (b-e). Source data for graphs are provided in Statistical Source Data Fig. 1 and unprocessed blots are shown in Unprocessed Blots Fig. 1.
Extended Data Fig. 2 TBK1, but not IKKε, mediates growth factor-stimulated mTORC1 activation.
a, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of freshly isolated lung cells from Tbk1WT-KrasLA2 and Tbk1cKO-KrasLA2 mice. b-e, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of control or Tbk1-knockdown H157 (b), H460 (c), or A549 (d,e) cells, stimulated with the indicated inducers. f, g, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of WT or Tbk1-deficient primary MEF cells stimulated by EGF (f) or essential amino acids (EAAs, g). h, Immunoblot analysis of the indicated proteins in the cytoplasmic extracts (Cyt Ext) or nuclear extracts (Nucl Ext) of control or Tbk1-knockdown A549 cells, stimulated with EGF. i, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of EGF-stimulated A549 cells stably infected with a control shRNA (shCtrl) or two different IKKe-specific shRNAs. Data are representative of three independent experiments. Unprocessed blots are shown in Unprocessed Blots Fig. 2.
Extended Data Fig. 3 TBK1 mediates mTORC1 activation by EGF and has cell type-specific functions in mTORC1 activation by TLR ligands.
a, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of A549 cells stably infected with a control shRNA (Ctrl) or two different Tbk1-specific shRNAs (D5 and D9), stimulated with Pam3Csk4. b-h, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of primary lung cells from WT and Tbk1cKO mice (b),Tbk1WT-KrasLA2 (WT-Kras) and Tbk1cKO-KrasLA2 (cKO-Kras) mice (c,d), bone marrow-derived macrophages (BMDM) from WT or Tbk1 myeloid cell-conditional KO (MKO) mice (e,f), or WT and Tbk1 KO primary MEFs (g,h), stimulated with the indicated TLR ligands. Data are representative of three independent experiments. Unprocessed blots are shown in Unprocessed Blots Fig. 3.
Extended Data Fig. 4 TBK1 is associated with mTORC1 via Raptor.
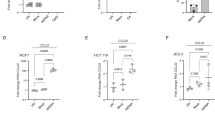
a, Schematic of Myc-BioID (Myc-tagged BirA R118G) and Myc-BioID-TBK1 constructs cloned into the pCLXSN(GFP) retroviral vector. b, Scatter plot showing several known TBK1-binding proteins and a novel TBK1-binding protein, Raptor (also called RPTOR) identified by BioID screening. c,d, Immunoblot analysis of Raptor and Rictor pulled down by TBK1 IP (c, upper) or mTOR and TBK1 pulled down by Rictor IP (d, upper), with protein expression being monitored by direct immunoblot assays (lower), using whole-cell lysates of HEK293 cells transfected with the indicated expression vectors. e, Co-IP analysis of endogenous TBK1-Raptor and TBK1-Rictor interactions in EGF-stimulated A549 cells (upper). IgG was used as a negative control for IP. f,g, Co-IP analysis of TBK1-mTORC1 interactions (upper) and direct immunoblot analyses of the indicated proteins (lower) in control or Tbk1-knockdown A549 cells stimulated with EGF. The Lysates were subjected to IP with antibody against TBK1 (f) or Raptor (g). Data are representative of three independent experiments. Unprocessed blots are shown in Unprocessed Blots Fig. 4.
Extended Data Fig. 5 TANK and NAP1 are dispensable for EGF-induced mTORC1 activation.
a-d, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of A549 cells stably infected with a control shRNA (shCtrl) or shRNAs specific for Tank (a), Nap1 (b), or Tbkbp1 (c,d), stimulated with the indicated inducers. e,f, Genotyping PCR (e) and immunoblot (f) analyses of Tbkbp1+/+Ccsp-Cre (WT), Tbkbp1+/flCcsp-Cre (Het), and Tbkbp1fl/flCcsp-Cre (cKO) mice, showing the flox and WT Tbk1 alleles and Ccsp-Cre (e) and the TBKBP1 protein expression (f). Data are representative of three independent experiments. Unprocessed blots are shown in Unprocessed Blots Fig. 5.
Extended Data Fig. 6 Role of TRAFs and PKC in TBK1 phosphorylation and activation.
a,b, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of EGF-stimulated A549 cells stably infected with a control shRNA (shCtrl) or two different shRNAs targeting Traf6 (a) or Traf3 (b). c,d, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of A549 cells stimulated with EGF in the presence of the PKC inhibitor GF109203X or solvent control DMSO. e, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in whole-cell lysates of control or PKCe-knockdown A549 cells, stimulated with EGF for the indicated time periods. f,g, Immunoblot analysis of T538-phosphorylated (p-) and total PKCq in the lysates of A549 cells stimulated with insulin (f) or FBS (g). h,i, immunoblot analysis of TBK1 and S172- or S716-phosphorylated (p-) TBK1 following HA IP from A549 cells stably transduced with HA-TBK1 and stimulated with either EGF (h) or Insulin (i). j, Immunoblot analysis of S172-phosphorylated (p-) and total TBK1 in whole-cell lysates of HEK293T cells transfected with the indicated expression vectors. k, CoIP assays to detect TBK1 dimerization based interaction of HA-TBK1 with Flag-TBK1 (upper) and immunoblot analysis of protein expression level in lysates (lower). l, Immunoblot analysis of the indicated phosphorylated (p-) and total proteins in LPS-stimulated control shRNA-transduced A549 cells or Tbk1 shRNA (D5)-transduced A549 cells that were reconstituted with and an empty vector or shRNA-resistant expression vectors encoding HA-tagged TBK1 wildtype (WT) or S716A. Data are representative of three independent experiments. Unprocessed blots are shown in Unprocessed Blots Fig. 6.
Extended Data Fig. 7 TBK1 deletion in lung epithelial cells promotes T cell activation.
a, Immunohistochemical staining of T cells using the indicated antibodies in lung sections of Tbk1WT-KrasLA2 (WT-Kras) and Tbk1cKO-KrasLA2 (cKO-Kras) mice. Scale bar, 100 mm. Data are presented as a representative image (upper) or summary graphs (lower, n = 7 mice per group). b,c, Flow cytometric analysis of the frequency of CD4 and CD8 T cells in lung cells (b) or IFNg+ effector T cells within lung CD4 or CD8 T cell populations (c). n = 5 mice per group. d-g, Experimental design (d), efficiency of T cell depletion in the lung and spleen (e), a representative image of lung tumours (f, scale bar, 100 mm), and summary graphs of lung weight, tumour numbers, and tumour size (g) of Tbk1WT-KrasLA2 and Tbk1cKO-KrasLA2 mice treated with anti-CD3 or an IgG isotype control (150 mg/mouse). n = 11 mice per group. Data are representative of three independent experiments, and bar graphs are presented as mean±s.d. values. Two-sided unpaired Student’s t-test (a,b,c,g). Source data for graphs are provided in Statistical Source Data Fig. 7.
Supplementary information
Supplementary Tables
Supplementary Table 1: summary of TBK1-binding proteins identified by BioID. Supplementary Table 2: primers used in quantitative RT–PCR and mouse genotyping PCR. Supplementary Table 3: list of antibodies used.
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 1
Unprocessed Blots
Source Data Fig. 2
Unprocessed Blots
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 3
Unprocessed Blots
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 4
Unprocessed Blots
Source Data Fig. 5
Unprocessed Blots
Source Data Fig. 6
Statistical Source Data
Source Data Fig. 6
Unprocessed Blots
Source Data Fig. 7
Statistical Source Data
Source Data Fig. 7
Unprocessed Blots
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 1
Unprocessed Blots
Source Data Extended Data Fig. 2
Unprocessed Blots
Source Data Extended Data Fig. 3
Unprocessed Blots
Source Data Extended Data Fig. 4
Unprocessed Blots
Source Data Extended Data Fig. 5
Unprocessed Blots
Source Data Extended Data Fig. 6
Unprocessed Blots
Source Data Extended Data Fig. 7
Statistical Source Data
Rights and permissions
About this article
Cite this article
Zhu, L., Li, Y., Xie, X. et al. TBKBP1 and TBK1 form a growth factor signalling axis mediating immunosuppression and tumourigenesis. Nat Cell Biol 21, 1604–1614 (2019). https://doi.org/10.1038/s41556-019-0429-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0429-8
This article is cited by
-
Metabolic reprogramming based on RNA sequencing of gemcitabine-resistant cells reveals the FASN gene as a therapeutic for bladder cancer
Journal of Translational Medicine (2024)
-
RAS signaling and immune cells: a sinister crosstalk in the tumor microenvironment
Journal of Translational Medicine (2023)
-
TRIM28 promotes the escape of gastric cancer cells from immune surveillance by increasing PD-L1 abundance
Signal Transduction and Targeted Therapy (2023)
-
Serine/threonine kinase TBK1 promotes cholangiocarcinoma progression via direct regulation of β-catenin
Oncogene (2023)
-
Exploring the mediation of DNA methylation across the epigenome between childhood adversity and First Episode of Psychosis—findings from the EU-GEI study
Molecular Psychiatry (2023)