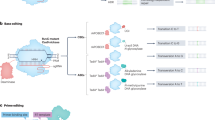
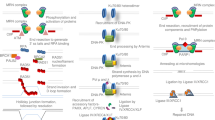
Abstract
Eukaryotic cells deploy overlapping repair pathways to resolve DNA damage. Advancements in genome editing take advantage of these pathways to produce permanent genetic changes. Despite recent improvements, genome editing can produce diverse outcomes that can introduce risks in clinical applications. Although homology-directed repair is attractive for its ability to encode precise edits, it is particularly difficult in human cells. Here we discuss the DNA repair pathways that underlie genome editing and strategies to favour various outcomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Cannan, W. J. & Pederson, D. S. Mechanisms and consequences of double-strand DNA break formation in chromatin. J. Cell. Physiol. 231, 3–14 (2016).
Ranjha, L., Howard, S. M. & Cejka, P. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma 127, 187–214 (2018).
Doudna, J. A. & Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014).
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S. & Gregory, P. D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Jinek, M. et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Jinek, M. et al. RNA-programmed genome editing in human cells. eLife 2, e00471 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR–Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Haber, J. E. A life investigating pathways that repair broken chromosomes. Annu. Rev. Genet. 50, 1–28 (2016).
Ward, J. F. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog. Nucleic Acid Res. Mol. Biol. 35, 95–125 (1988).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
McConnell Smith, A. et al. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc. Natl Acad. Sci. 106, 5099–5104 (2009).
Davis, L. & Maizels, N. DNA nicks promote efficient and safe targeted gene correction. PLoS One 6, e23981 (2011).
Davis, L. & Maizels, N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. 111, E924–E932 (2014).
Davis, L. & Maizels, N. Two distinct pathways support gene correction by single-stranded donors at DNA nicks. Cell Rep. 17, 1872–1881 (2016).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Schatz, D. G. & Ji, Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11, 251–263 (2011).
Baudat, F., Imai, Y. & de Massy, B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 14, 794–806 (2013).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015).
Mali, P. et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 (2013).
Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L. & Corn, J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344 (2016).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Knight, S. C. et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science 350, 823–826 (2015).
Clarke, R. et al. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol. Cell 71, 42–55.e8 (2018).
DiCarlo, J. E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013).
Mao, Z., Bozzella, M., Seluanov, A. & Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 7, 1765–1771 (2008).
Chang, H. H. Y., Pannunzio, N. R., Adachi, N. & Lieber, M. R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017).
Mjelle, R. et al. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair (Amst.) 30, 53–67 (2015).
Ren, K. & Peña de Ortiz, S. Non-homologous DNA end joining in the mature rat brain. J. Neurochem. 80, 949–959 (2002).
Bae, S., Kweon, J., Kim, H. S. & Kim, J.-S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11, 705–706 (2014).
Shen, M. W. et al. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018).
Griffith, A. J., Blier, P. R., Mimori, T. & Hardin, J. A. Ku polypeptides synthesized in vitro assemble into complexes which recognize ends of double-stranded DNA. J. Biol. Chem. 267, 331–338 (1992).
Spagnolo, L., Rivera-Calzada, A., Pearl, L. H. & Llorca, O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell 22, 511–519 (2006).
Davis, A. J., Chen, B. P. C. & Chen, D. J. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst.) 17, 21–29 (2014).
Yang, S. et al. The SOSS1 single‐stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 32, 126–139 (2013).
Mimitou, E. P. & Symington, L. S. Ku prevents Exo1 and Sgs1‐dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369 (2010).
Shim, E. Y. et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380 (2010).
Mari, P.-O. et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. 103, 18597–18602 (2006).
Bryans, M., Valenzano, M. C. & Stamato, T. D. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat. Res. 433, 53–58 (1999).
Yano, K. & Chen, D. J. Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle 7, 1321–1325 (2008).
Chang, H. H. Y., Watanabe, G. & Lieber, M. R. Unifying the DNA end-processing roles of the artemis nuclease: Ku-dependent artemis resection at blunt DNA ends. J. Biol. Chem. 290, 24036–24050 (2015).
Mahaney, B. L., Meek, K. & Lees-Miller, S. P. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417, 639–650 (2009).
Riesenberg, S. & Maricic, T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun. 9, 2164 (2018).
Ninomiya, Y., Suzuki, K., Ishii, C. & Inoue, H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101, 12248–12253 (2004).
da Silva Ferreira, M. E. et al. The akuB KU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in. Aspergillus fumigatus. Eukaryot. Cell 5, 207–211 (2006).
Fattah, F. J., Lichter, N. F., Fattah, K. R., Oh, S. & Hendrickson, E. A. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc. Natl. Acad. Sci. 105, 8703–8708 (2008).
Srivastava, M. et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 151, 1474–1487 (2012).
Chu, V. T. et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015).
Beumer, K. J. et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. 105, 19821–19826 (2008).
Maruyama, T. et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 33, 538–542 (2015).
Robert, F., Barbeau, M., Éthier, S., Dostie, J. & Pelletier, J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 7, 93 (2015).
Singh, P., Schimenti, J. C. & Bolcun-Filas, E. A mouse geneticist’s practical guide to CRISPR applications. Genetics 199, 1–15 (2015).
Hu, Z. et al. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR-Cas9 and ssODN in human cancer cells. Cell Biosci. 8, 12 (2018).
Song, J. et al. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 7, 10548 (2016).
Zhang, J.-P. et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 18, 35 (2017).
Gutschner, T., Haemmerle, M., Genovese, G., Draetta, G. F. & Chin, L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 14, 1555–1566 (2016).
Yang, D. et al. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 6, 21264 (2016).
Pinder, J., Salsman, J. & Dellaire, G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 43, 9379–9392 (2015).
Greco, G. E. et al. SCR7 is neither a selective nor a potent inhibitor of human DNA ligase IV. DNA Repair (Amst.) 43, 18–23 (2016).
Truong, L. N. et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl Acad. Sci. 110, 7720–7725 (2013).
Yan, C. T. et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007).
Boboila, C. et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 207, 417–427 (2010).
Myler, L. R. et al. Single-molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. Mol. Cell 67, 891–898.e4 (2017).
Anand, R., Ranjha, L., Cannavo, E. & Cejka, P. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950 (2016).
Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007).
Shibata, A. et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 (2014).
Garcia, V., Phelps, S. E. L., Gray, S. & Neale, M. J. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479, 241–244 (2011).
Zhou, Y., Caron, P., Legube, G. & Paull, T. T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 42, e19 (2014).
Paull, T. T. & Gellert, M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979 (1998).
Nakade, S. et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 5, 5560 (2014).
Ahmad, A. et al. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol. Cell. Biol. 28, 5082–5092 (2008).
Kent, T., Chandramouly, G., McDevitt, S. M., Ozdemir, A. Y. & Pomerantz, R. T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 (2015).
Liang, L. et al. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 36, 3297–3310 (2008).
Audebert, M., Salles, B. & Calsou, P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279, 55117–55126 (2004).
Wang, M. et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170–6182 (2006).
Haince, J.-F. et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 (2008).
Mateos-Gomez, P. A. et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015).
Iyer, S. et al. Precise therapeutic gene correction by a simple nuclease-induced double-stranded break. Nature 568, 561–565 (2019).
Mateos-Gomez, P. A. et al. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat. Struct. Mol. Biol. 24, 1116–1123 (2017).
Dutta, A. et al. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 45, 2585–2599 (2017).
Allen, F. et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 37, 64–72 (2018).
Jiang, F. et al. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016).
Shou, J., Li, J., Liu, Y. & Wu, Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol. Cell 71, 498–509.e4 (2018).
Richardson, C. D. et al. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 50, 1132–1139 (2018).
Liang, F., Han, M., Romanienko, P. J. & Jasin, M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. 95, 5172–5177 (1998).
Zhu, Z., Chung, W.-H., Shim, E. Y., Lee, S. E. & Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008).
Chen, R. & Wold, M. S. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays 36, 1156–1161 (2014).
Maréchal, A. & Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 5, a012716 (2013).
Ihry, R. J. et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Nakanishi, K. et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. USA 102, 1110–1115 (2005).
Ceccaldi, R., Sarangi, P. & D’Andrea, A. D. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 17, 337–349 (2016).
Roques, C. et al. MRE11–RAD50–NBS1 is a critical regulator of FANCD2 stability and function during DNA double‐strand break repair. EMBO J. 28, 2400–2413 (2009).
Unno, J. et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 7, 1039–1047 (2014).
Howard, S. M., Yanez, D. A. & Stark, J. M. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 11, e1004943 (2015).
Benitez, A. et al. FANCA promotes DNA double-strand break repair by catalyzing single-strand annealing and strand exchange. Mol. Cell 71, 621–628.e4 (2018).
Sugiyama, T., Zaitseva, E. M. & Kowalczykowski, S. C. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 272, 7940–7945 (1997).
Renkawitz, J., Lademann, C. A., Kalocsay, M. & Jentsch, S. Monitoring homology search during DNA double-strand break repair in vivo. Mol. Cell 50, 261–272 (2013).
Sugawara, N., Ira, G. & Haber, J. E. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20, 5300–5309 (2000).
Chun, J., Buechelmaier, E. S. & Powell, S. N. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 33, 387–395 (2013).
Zhang, S. et al. Structural basis for the functional role of the Shu complex in homologous recombination. Nucleic Acids Res. 45, 13068–13079 (2017).
Liu, J., Doty, T., Gibson, B. & Heyer, W.-D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 (2010).
McVey, M., Khodaverdian, V. Y., Meyer, D., Cerqueira, P. G. & Heyer, W.-D. Eukaryotic DNA polymerases in homologous recombination. Annu. Rev. Genet. 50, 393–421 (2016).
Verma, P. & Greenberg, R. A. Noncanonical views of homology-directed DNA repair. Genes Dev. 30, 1138–1154 (2016).
Goetz, J. D.-M., Motycka, T. A., Han, M., Jasin, M. & Tomkinson, A. E. Reduced repair of DNA double-strand breaks by homologous recombination in a DNA ligase I-deficient human cell line. DNA Repair (Amst.) 4, 649–654 (2005).
Bentley, D. et al. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat. Genet. 13, 489–491 (1996).
Bugreev, D. V., Yu, X., Egelman, E. H. & Mazin, A. V. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 21, 3085–3094 (2007).
Hu, Y. et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 (2007).
Fugger, K. et al. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 186, 655–663 (2009).
Simandlova, J. et al. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 288, 34168–34180 (2013).
Chu, W. K. et al. FBH1 influences DNA replication fork stability and homologous recombination through ubiquitylation of RAD51. Nat. Commun. 6, 5931 (2015).
Jayathilaka, K. et al. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. 105, 15848–15853 (2008).
Huang, F., Mazina, O. M., Zentner, I. J., Cocklin, S. & Mazin, A. V. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. J. Med. Chem. 55, 3011–3020 (2012).
Ren, C., Yan, Q. & Zhang, Z. Minimum length of direct repeat sequences required for efficient homologous recombination induced by zinc finger nuclease in yeast. Mol. Biol. Rep. 41, 6939–6948 (2014).
Liskay, R. M., Letsou, A. & Stachelek, J. L. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics 115, 161–167 (1987).
Mortensen, U. H., Bendixen, C., Sunjevaric, I. & Rothstein, R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. 93, 10729–10734 (1996).
Shinohara, A., Shinohara, M., Ohta, T., Matsuda, S. & Ogawa, T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells 3, 145–156 (1998).
Rothenberg, E., Grimme, J. M., Spies, M. & Ha, T. Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. 105, 20274–20279 (2008).
Grimme, J. M. et al. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 38, 2917–2930 (2010).
Han, J. et al. BRCA2 antagonizes classical and alternative nonhomologous end-joining to prevent gross genomic instability. Nat. Commun. 8, 1470 (2017).
Ma, C. J., Kwon, Y., Sung, P. & Greene, E. C. Human RAD52 interactions with replication protein A and the RAD51 presynaptic complex. J. Biol. Chem. 292, 11702–11713 (2017).
Feng, Z. et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. 108, 686–691 (2011).
Li, X. et al. Efficient SSA-mediated precise genome editing using CRISPR/Cas9. FEBS J. 285, 3362–3375 (2018).
German, J. Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 72, 393–406 (1993).
Aylon, Y., Liefshitz, B. & Kupiec, M. The CDK regulates repair of double‐strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875 (2004).
Ira, G. et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017 (2004).
Yun, M. H. & Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 (2009).
Buis, J., Stoneham, T., Spehalski, E. & Ferguson, D. O. Mre11 regulates CtIP-dependent double-strand break repair by interaction with CDK2. Nat. Struct. Mol. Biol. 19, 246–252 (2012).
Peterson, S. E. et al. Cdk1 uncouples CtIP-dependent resection and Rad51 filament formation during M-phase double-strand break repair. J. Cell Biol. 194, 705–720 (2011).
Charpentier, M. et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 9, 1133 (2018).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Escribano-Díaz, C. et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013).
Gupta, R. et al. DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173, 972–988.e23 (2018).
Noordermeer, S. M. et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018).
Orthwein, A. et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 (2015).
Paulsen, B. S. et al. Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat. Biomed. Eng. 1, 878–888 (2017).
Canny, M. D. et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat. Biotechnol. 36, 95–102 (2018).
Ye, L. et al. Programmable DNA repair with CRISPRa/i enhanced homology-directed repair efficiency with a single Cas9. Cell Discov. 4, 46 (2018).
Arnoult, N. et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 (2017).
Hung, P. J. et al. MRI is a DNA damage response adaptor during classical non-homologous end joining. Mol. Cell 71, 332–342.e8 (2018).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
Lomova, A. et al. Improving gene editing outcomes in human hematopoietic stem and progenitor cells by temporal control of DNA repair. Stem Cells 37, 284–294 (2018).
Storici, F., Snipe, J. R., Chan, G. K., Gordenin, D. A. & Resnick, M. A. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol. Cell. Biol. 26, 7645–7657 (2006).
Chen, F. et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755 (2011).
Yang, L. et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 41, 9049–9061 (2013).
DeWitt, M. A. et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci. Transl. Med. 8, 360ra134 (2016).
Kan, Y., Ruis, B., Takasugi, T. & Hendrickson, E. A. Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 27, 1099–1111 (2017).
Bothmer, A. et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 8, 13905 (2017).
Shao, S. et al. Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cell Biol. 92, 43–52 (2017).
Liang, X., Potter, J., Kumar, S., Ravinder, N. & Chesnut, J. D. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J. Biotechnol. 241, 136–146 (2017).
Renkawitz, J., Lademann, C. A. & Jentsch, S. Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 15, 369–383 (2014).
Ünal, E. et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 (2004).
Ström, L., Lindroos, H. B., Shirahige, K. & Sjögren, C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 (2004).
Miné-Hattab, J. & Rothstein, R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517 (2012).
Schrank, B. R. et al. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66 (2018).
Savic, N. et al. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife 7, e33761 (2018).
Aird, E. J., Lovendahl, K. N., St Martin, A., Harris, R. S. & Gordon, W. R. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 1, 54 (2018).
Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 24, 374–387 (2014).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature https://doi.org/10.1038/s41586-019-1711-4 (2019)
Kass, E. M., Lim, P. X., Helgadottir, H. R., Moynahan, M. E. & Jasin, M. Robust homology-directed repair within mouse mammary tissue is not specifically affected by Brca2 mutation. Nat. Commun. 7, 13241 (2016).
Stork, C. T. et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife 5, e17548 (2016).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
van Overbeek, M. et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 63, 633–646 (2016).
Taheri-Ghahfarokhi, A. et al. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 46, 8417–8434 (2018).
Wang, K. et al. Efficient generation of orthologous point mutations in pigs via CRISPR-assisted ssODN-mediated homology-directed repair. Mol. Ther. Nucleic Acids 5, e396 (2016).
Yu, C. et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16, 142–147 (2015).
Acknowledgements
This work was supported by the Li Ka Shing Foundation (J.E.C., C.D.R. and C.D.Y.), the NIH New Innovator Program (DP2 HL141006, J.E.C.), the Heritage Medical Research Institute (C.D.R. and C.D.Y.), and the Fanconi Anemia Research Foundation (C.D.R. and C.D.Y.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yeh, C.D., Richardson, C.D. & Corn, J.E. Advances in genome editing through control of DNA repair pathways. Nat Cell Biol 21, 1468–1478 (2019). https://doi.org/10.1038/s41556-019-0425-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0425-z
This article is cited by
-
HiHo-AID2: boosting homozygous knock-in efficiency enables robust generation of human auxin-inducible degron cells
Genome Biology (2024)
-
Improving CRISPR–Cas9 directed faithful transgene integration outcomes by reducing unwanted random DNA integration
Journal of Biomedical Science (2024)
-
Enabling regulatory policy globally will promote realization of the potential of animal biotechnology
CABI Agriculture and Bioscience (2024)
-
Dbf4-dependent kinase promotes cell cycle controlled resection of DNA double-strand breaks and repair by homologous recombination
Nature Communications (2024)
-
Transient inhibition of 53BP1 increases the frequency of targeted integration in human hematopoietic stem and progenitor cells
Nature Communications (2024)