Abstract
With ageing, intrinsic haematopoietic stem cell (HSC) activity decreases, resulting in impaired tissue homeostasis, reduced engraftment following transplantation and increased susceptibility to diseases. However, whether ageing also affects the HSC niche, and thereby impairs its capacity to support HSC function, is still widely debated. Here, by using in-vivo long-term label-retention assays we demonstrate that aged label-retaining HSCs, which are, in old mice, the most quiescent HSC subpopulation with the highest regenerative capacity and cellular polarity, reside predominantly in perisinusoidal niches. Furthermore, we demonstrate that sinusoidal niches are uniquely preserved in shape, morphology and number on ageing. Finally, we show that myeloablative chemotherapy can selectively disrupt aged sinusoidal niches in the long term, which is linked to the lack of recovery of endothelial Jag2 at sinusoids. Overall, our data characterize the functional alterations of the aged HSC niche and unveil that perisinusoidal niches are uniquely preserved and thereby protect HSCs from ageing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq data that support the findings of this study have been deposited in the gene expression omnibus under accession code GSE129726. Previously published sequencing data that were re-analysed here are available under ArrayExpress E-MTAB-454723. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Change history
22 November 2019
In the version of this article originally published, the files for the Supplementary Tables and the main figure Source Data were linked incorrectly. This has now been corrected.
References
Kunisaki, Y. et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature 502, 637–643 (2013).
Ding, L., Saunders, T. L., Enikolopov, G. & Morrison, S. J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012).
Acar, M. et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015).
Itkin, T. et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature 532, 323–328 (2016).
Chen, J. Y. et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227 (2016).
Calvi, L. M. et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425, 841–846 (2003).
Guidi, N. et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J. 36, 840–853 (2017).
Heazlewood, S. Y. et al. Megakaryocytes co-localise with hemopoietic stem cells and release cytokines that up-regulate stem cell proliferation. Stem Cell Res. 11, 782–792 (2013).
Zhao, M. et al. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med. 20, 1321–1326 (2014).
Bruns, I. et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nat. Med. 20, 1315–1320 (2014).
Mendez-Ferrer, S. et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834 (2010).
Haylock, D. N. et al. Hemopoietic stem cells with higher hemopoietic potential reside at the bone marrow endosteum. Stem Cells 25, 1062–1069 (2007).
Akunuru, S. & Geiger, H. Aging, Clonality, and Rejuvenation of Hematopoietic Stem Cells. Trends Mol. Med. 22, 701–712 (2016).
Geiger, H., de Haan, G. & Florian, M. C. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 13, 376–389 (2013).
Morrison, S. J., Wandycz, A. M., Akashi, K., Globerson, A. & Weissman, I. L. The aging of hematopoietic stem cells. Nat. Med. 2, 1011–1016 (1996).
Rossi, D. J. et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad. Sci. USA 102, 9194–9199 (2005).
Kamminga, L. M. et al. Impaired hematopoietic stem cell functioning after serial transplantation and during normal aging. Stem Cells 23, 82–92 (2005).
Beerman, I., Maloney, W. J., Weissmann, I. L. & Rossi, D. J. Stem cells and the aging hematopoietic system. Curr. Opin. Immunol. 22, 500–506 (2010).
Maryanovich, M. et al. Adrenergic nerve degeneration in bone marrow drives aging of the hematopoietic stem cell niche. Nat. Med. 24, 782–791 (2018).
Bockamp, E. et al. Tetracycline-controlled transgenic targeting from the SCL locus directs conditional expression to erythrocytes, megakaryocytes, granulocytes, and c-kit-expressing lineage-negative hematopoietic cells. Blood 108, 1533–1541 (2006).
Wilson, A. et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008).
Walter, D. et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520, 549–552 (2015).
Cabezas-Wallscheid, N. et al. Vitamin a-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell 169, 807–823 (2017).
Essers, M. A. et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 458, 904–908 (2009).
Bernitz, J. M., Kim, H. S., MacArthur, B., Sieburg, H. & Moore, K. Hematopoietic stem cells count and remember self-renewal divisions. Cell 167, 1296–1309 (2016).
Waskow, C. et al. Hematopoietic stem cell transplantation without irradiation. Nat. Methods 6, 267–269 (2009).
Florian, M. C. et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature 503, 392–396 (2013).
Florian, M. C. et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 10, 520–530 (2012).
Grigoryan, A. et al. LaminA/C regulates epigenetic and chromatin architecture changes upon aging of hematopoietic stem cells. Genome Biol. 19, 189 (2018).
Florian, M. C. et al. Aging alters the epigenetic asymmetry of HSC division. PLoS Biol. 16, e2003389 (2018).
Maryanovich, M., Takeishi, S. & Frenette, P. S. Neural regulation of bone and bone marrow. Cold Spring Harb. Perspect. Med. 4, a031344 (2018).
Kusumbe, A. P. et al. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature 532, 380–384 (2016).
Spencer, J. A. et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature 508, 269–273 (2014).
Zhang, Y. et al. CXCR4/CXCL12 axis counteracts hematopoietic stem cell exhaustion through selective protection against oxidative stress. Sci. Rep. 6, 37827 (2016).
Sugiyama, T., Kohara, H., Noda, M. & Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25, 977–988 (2006).
Broxmeyer, H. E. et al. DEK regulates hematopoietic stem engraftment and progenitor cell proliferation. Stem Cells Dev. 21, 1449–1454 (2012).
Solaimani Kartalaei, P. et al. Whole-transcriptome analysis of endothelial to hematopoietic stem cell transition reveals a requirement for Gpr56 in HSC generation. J. Exp. Med. 212, 93–106 (2015).
Holmfeldt, P. et al. Functional screen identifies regulators of murine hematopoietic stem cell repopulation. J. Exp. Med. 213, 433–449 (2016).
Borggrefe, T. & Oswald, F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 66, 1631–1646 (2009).
Kojika, S. & Griffin, J. D. Notch receptors and hematopoiesis. Exp. Hematol. 29, 1041–1052 (2001).
Guiu, J. et al. Hes repressors are essential regulators of hematopoietic stem cell development downstream of Notch signaling. J. Exp. Med. 210, 71–84 (2013).
Shimizu, K. et al. Binding of Delta1, Jagged1, and Jagged2 to Notch2 rapidly induces cleavage, nuclear translocation, and hyperphosphorylation of Notch2. Mol. Cell. Biol. 20, 6913–6922 (2000).
Kopp, H. G., Hooper, A. T., Avecilla, S. T. & Rafii, S. Functional heterogeneity of the bone marrow vascular niche. Ann. N. Y. Acad. Sci. 1176, 47–54 (2009).
Kopp, H. G. et al. Tie2 activation contributes to hemangiogenic regeneration after myelosuppression. Blood 106, 505–513 (2005).
Zhou, B. O. et al. Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nat. Cell Biol. 19, 891–903 (2017).
Baryawno, N., Severe, N. & Scadden, D. T. Hematopoiesis: reconciling historic controversies about the niche. Cell Stem Cell 20, 590–592 (2017).
Maillard, I. et al. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell 2, 356–366 (2008).
Guo, P. et al. Endothelial jagged-2 sustains hematopoietic stem and progenitor reconstitution after myelosuppression. J. Clin. Invest. 127, 4242–4256 (2017).
Mazurier, F. et al. A novel immunodeficient mouse model–RAG2 x common cytokine receptor gamma chain double mutants–requiring exogenous cytokine administration for human hematopoietic stem cell engraftment. J. Interferon Cytokine Res. 19, 533–541 (1999).
Luche, H., Weber, O., Nageswara Rao, T., Blum, C. & Fehling, H. J. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37, 43–53 (2007).
Schwenk, F., Baron, U. & Rajewsky, K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–5081 (1995).
Lis, R. et al. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature 545, 439–445 (2017).
Elyaman, W. et al. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity 36, 623–634 (2012).
Angerer, P. et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016).
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Brennecke, P. et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Scialdone, A. et al. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods 85, 54–61 (2015).
Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016).
Acknowledgements
We thank J. A. Cancelas (Cincinnati Children Hospital Medical Center) for advice and critical reading of the manuscript. We thank H. J. Fehling (Institute of Immunology, Ulm University) for donating AcRFP and AcYFP mice and Y. Zheng (Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA) for donating AcCFP mice. We thank the core facilities at Ulm University, A. Rück (Microscope Facility), for support with confocal and two-photon microscopy, as well as the Cell Sorting Facility and the Tierforschungszentrum (Animal Facility). Work in the laboratory of M.C.F. was supported by an Emmy Noether Grant (DFG) (grant nos. SFB1074 and FOR2674) to M.C.F. M.S. was supported by a PhD Degree Scholarship of the Avicenna-Studienwerk programme.
Author information
Authors and Affiliations
Contributions
M.S. and M.C.F. performed and analysed the laboratory experiments. M.S. performed all histological experiments and microscopy analysis. J.P. conceived the computational model with assistance of W.d.B. and performed the bioinformatic analysis of scRNA-seq datasets with support from R.K. K.S. and G.M. assisted in transplantation procedures, bleeding, supported in cell sorting and flow analysis procedures. A.V. performed single cell RNA-seq sample preparation, supported in cell sorting and flow analysis procedures and performed in vitro co-culture experiments. V.S. supervised the mouse work and took care of breeding, ageing and preparation of the mice used for experiments. M.S. and M.C.F. designed and interpreted the experiments. J.-P.M. prepared scRNA-seq libraries from endothelial cells. N.C.-W. supported the scRNA sequencing. N.C.-W., A.T., M.D.M. and R.B. supported all the experiments involving SCL-tTAxH2B-GFP double heterozygous mice. M.A.M. developed the deep learning algorithm. H.G. helped interpret the experiments and write the manuscript. S.M.-F. supported with Nestin-GFP mice and assisted in interpreting experiments. M.S., J.P. and M.C.F. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
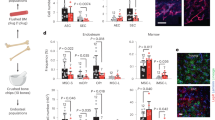
Extended Data Fig. 1 Aged LR-HSC pool expand less than aged nLR-HSCs and are functional long-term stem cells.
a, Representative FACS dot plots of the gating strategy for LR-HSCs (Lin−c-Kit+ Sca-1+ Flk2−CD34−CD48−CD150+ H2B-GFP+) of young and aged not DOX treated H2B-GFP, and young and aged DOX treated SCL-tTAxH2B-GFP lineage-depleted BM cells and schematic representation of the experimental setups. b, Frequency of different HSPC subpopulations, LR-HSCs and nLR-HSCs among BM cells in young and old SCL-tTAxH2B-GFP mice after 2 and 18-months of DOX chase, respectively. c, Percentages of young and old LR-HSCs in BM cells in DOX treated SCL-tTAxH2B-GFP mice. d, Similar frequency of old LSK CD34−Flk2−HSCs and old LSK CD34−Flk2−CD48−CD150+ HSCs among BM cells. e, Fold expansion of different HSPC subpopulations, LR-HSCs and nLR-HSCs with ageing. f, Experimental setup of single aged LR-HSC transplantation in Rag2-/-γc-/-KitW/Wv recipient mouse and gating strategy. g, Pie chart showing percentage of functional long-term HSCs among single transplanted aged LR-HSCs. h, Blood chimerism kinetics of overall engrafted donor-derived cells and of each donor-derived lineage (B cells, T cells and myeloid cells) with single donor old LR-HSCs. The experiments in a were repeated four independent times with similar results. Data in b-d represent mean ± s.e.m. In b-e, n = 4 young and 5 aged mice. In g, h n = 7 recipient mice. The statistical significance was assessed by two-tailed unpaired t-tests. * p < 0.05. See also Statistical Source Data Extended Data Fig. 1.
Extended Data Fig. 2 Aged LR-HSCs show higher regenerative capacity and are polar.
a, Scheme of the transplantation. b, Representative blood chimerism kinetics of overall engrafted donor-derived cells and of each donor-derived lineage (B cells, T cells and myeloid cells) during primary and secondary transplants of one recipient mouse transplanted with donor aLR-HSCs and one with donor anLR-HSCs. c, Frequency of old nLR-HSCs and old LR-HSCs donor contribution to total white blood cells (WBCs) in PB of Rag2-/-γc-/-KitW/Wv recipient mice (n = 8 mice for nLR-HSCs, n = 9 mice for LR-HSCs). d, Frequency of aged LR- and nLR‐HSCs in BM among donor-derived LSK cells in Rag2-/-γc-/-KitW/Wv recipient mice. (n = 5 mice per group) e, Percentage of B cells (B220+) and T cells (Cd3+) in PB. Rag2-/-γc-/-KitW/Wv recipient mice are devoid of B and T cells per definition26,49] and all B220+ and Cd3+ cells among total WBC where donor-derived. Myeloid cells (Gr1+, Mac1+ and Gr1+Mac1+) are gated from total WBC and plotted as percentage of donor derived myeloid cells among total myeloid cells. Cartoon schemes on top of each graph depicts the gating strategy (n = 8 mice for nLR-HSCs, n = 9 mice for LR-HSCs). f, Representative single-cell IF images showing tubulin (green) and Cdc42 (red), in old LR-HSCs and old nLR-HSCs. Nuclei, DAPI (blue). g, Representative 3D reconstructed single-cell IF confocal images showing H4K16ac (magenta) in old LR-HSCs (polar) and old nLR-HSCs (apolar). Nuclei, DAPI (blue). h, i, Percentage of polar cells (h) in old LR-HSCs and nLR-HSCs (n = 3 mice for Cdc42, n = 4 for H4K16ac, n = 7 for Tubulin) and in (i) young LR-HSCs and nLR-HSCs (n = 3 mice for Cdc42, AcH4K16; n = 5 mice for Tubulin). Data in h, i represent mean ± s.e.m, in c-e data represent the 25th-75th percentiles, minimum, maximum, median (bar) and mean (cross). The statistical significance was assessed by two-tailed unpaired t-tests for all data beside in e by two-way ANOVA-test. In e p 0.0397 is related to overall lymphoid cells (B220+ and Cd3+ data together) between aLR-HSC and anLR-HSC samples. As for myeloid cells p = 0.09. The statistical details for c-e are available in Source Data Extended Data Fig. 2.
Extended Data Fig. 3 Young LR-HSCs are located in central and endosteal BM.
a, Representative stacked whole-mount images of young SCL-tTAxH2B-GFP femoral BM showing LR cells (green) and vasculature (blue). b, 3D reconstruction of confocal whole-mount images of young SCL-tTAxH2B-GFP femoral BM showing a CD150+ H2B-GFP+ LR cell in proximity to arterial vasculature (arrowheads) in the endosteal area. c, d, Distinction of sinusoids and arterioles in (c) FABP4 (red) in situ stained and (d) CD31/CD144 (red) i.v. stained BM: arterioles (yellow arrowheads) have small diameter with continuous staining (continuous basal lamina) and have parallel orientation to the long axis of the bone, sinusoids (blue arrowheads) present relatively larger diameter with spotted staining (fenestrated basal lamina) and are mostly transverse to the long axis. (c) Nuclei, DAPI (blue). e, f, Colocalization of (e) sinusoids and (f) arterioles/arteries in FABP4 (green) in situ stained and CD31/CD144 (red) i.v. stained BM. Experiments in a-c, e, f were repeated three independent times with similar results and in d at least 9 independent times with similar results. Scale bars are 100 μm (a); 20 μm (b-f).
Extended Data Fig. 4 HSC proximity to selected niche cell types is altered upon ageing.
a, Distance between young and old HSCs to the nearest endosteum (192 young HSCs from 36 areas and 432 old HSCs from 17 areas, n = 3 mice per group), b, Confocal images of young and old whole-mount WT femoral BM showing HSCs (arrows) and MKs. c, Percentage of young and old HSCs in direct proximity to MKs ( < 10 µm; 177 young HSCs from 34 areas and 163 old HSCs from 10 areas, n = 3 mice per group). d, Mean distance of young and old HSCs to the nearest MK (n = 177 young HSCs from 34 areas and n = 163 old HSCs from 10 areas, 3 mice per group). e, Distance between young and old HSCs to the nearest MK (177 young HSCs from 34 areas and 163 old HSCs from 10 areas, n = 3 mice per group). f, Confocal whole-mount images of young and old sternal Nes-GFP mice BM. Arrows (yellow) show HSCs. Nuclei, DAPI (blue). g, Percentage of young and old HSCs in direct contact to Nes-GFPhigh cells ( < 1 µm; 137 young HSCs from 9 areas and 275 old HSCs from 12, n = 3 mice per group. h, Mean distance of young and old HSCs to the nearest Nes-GFPhigh cell (n = 137 young HSCs from 9 areas and n = 275 old HSCs from 12 areas, 3 mice per group). i, Mean distance of young and old HSCs to the nearest Nes-GFPlow cell (n = 94 young HSCs from 7 areas and n = 135 old HSCs from 7 areas, 3 mice per group). j, k, Distance between young and old HSCs to the nearest (j) Nes-GFPhigh cell (137 young HSCs from 9 areas and 275 old HSCs from 12 areas. n = 3 mice per group) and (k) Nes-GFPlow cell, (94 young HSCs from 7 areas and 135 old HSCs from 7 areas. n = 3 mice per group). The experiments in b, f were repeated three independent times with similar results. Data represent mean ± s.e.m. In d, h, i the statistical significance was assessed by two-tailed Mann-Whitney-test, in a, e, j, k the statistical significance was assessed by two-way-ANOVA-test and in c, g by two-tailed unpaired t-tests. Scale bars, 20 µm (b); 40 μm (f). See also Source Data Extended Data Fig. 4.
Extended Data Fig. 5 HSC proximity to LepR+ cells and HSC Ki67 + frequency are not altered upon ageing.
a, Whole-mount confocal images revealing the overlap of perivascular LepR+ cells (red) and Nes-GFPlow cells (green) surrounding sinusoidal vessels (blue) in central BM of Nes-GFP mice. b, Percentage of young and old HSCs adjacent to perivascular LepR+ cells ( < 10 µm; 96 young HSCs from 12 areas and 147 old HSCs from 12 areas, n = 3 femurs and two mice per group). c, Mean distance of young and old HSCs to the nearest perivascular LepR+ cells ( < 10 µm; n = 96 young HSCs from 12 areas and n = 147 old HSCs from 12 areas, 3 femurs and two mice per group). d, Percentages of young and old HSCs to the nearest perivascular LepR+ cell. (96 young HSCs from 12 areas and 147 old HSCs from 12 areas, n = 3 femurs and two mice per group). e, Percentage of young and old HSCs in proximity to BM vasculature ( < 10 µm; 193 young HSCs and 397 old HSCs, n = 3 mice per group). f, Mean distance of young and old HSCs to the nearest vasculature (n = 193 young HSCs and n = 397 old HSCs, 3 mice per group). g, Percentages of young and old HSCs to the nearest vasculature (193 young HSCs and 397 old HSCs, n = 3 mice per group). h, Confocal images of WT old femoral BM. Arrow (yellow) shows Ki-67+ (green) HSC. i, Percentage of Ki-67+ and Ki-67− HSCs (CD150+ CD41− CD48− Lin−) present in femoral whole-mount images in BM of WT young and old mice (n = 43 young and n = 140 old HSCs, 3 mice per group). j, 2-photon microscopy images showing Nes-GFP+ cells (green) at the endosteum of young and aged Nes-GFP mouse BM. Bone (collagen, white) depiction was generated with second-harmonic generation microscopy (1 unit = 13.53 µm). Experiments in a were repeated two in h, j three independent times with similar results. Data represent mean ± s.e.m. In c, f the statistical significance was assessed by two-tailed Mann-Whitney-test, in d, g the statistical significance was assessed by two-way-ANOVA-test and in b, e by two-tailed unpaired t-tests. Scale bars, 20 µm (a, h). See also Source Data Extended Data Fig. 5.
Extended Data Fig. 6 HSC proximity to selected niche cell types is not random.
a, Representative stacked whole-mount images of WT femoral BM showing CD41+ (yellow) MKs and vasculature (red). b, Confocal whole-mount image of WT femoral BM showing perisinusoidal CD41+ (yellow) MKs around sinusoids (red). c, Representative 3D reconstruction for volume occupancy measurements of CD31+ CD144+ ECs in femoral whole-mount images. d, Epiphyseal/metaphyseal and diaphyseal CD31+ CD144+ ECs volume occupancy in young and old long bone BM (upper graph: n = 3 areas per group, from 3 mice per group; lower graph: young n = 9 and old n = 13 areas, from 3 mice per group). The experiments in a-c were repeated two independent times with similar results. In d data represent mean ± s.e.m and the statistical significance was assessed by two-tailed unpaired t-tests. Scale bars, 100 μm (a); 20 μm (b). e-g, Observed (histology) and expected (in silico) distances of HSCs from endosteum (e), Nes-GFPlow cells (f) or MKs (g) in young and aged mice. Statistical differences indicate a non-random distribution of HSCs. h-j, Comparison of the distances of HSCs from endosteum (h), Nes-GFPlow cells (i) or MKs (j) in young and aged mice according histology (observed young vs observed aged) and in silico (expected young vs expected aged) dataset. For e-j two-sample Kolmogorov–Smirnov test was used to analyse significance. Alpha levels were selected to control for sample sizes. Observed distance from Nes-GFPlow cells: aged: n = 135 measurements from two maps; young: n = 94 measurements from two maps. Observed distance from endosteum: aged: n = 432 measurements from 3 maps; young: n = 192 measurements from 3 maps. Observed distance from MKs: aged: n = 163 measurements from 3 maps; young: n = 123 measurements from two maps. Expected distance from Nes-GFPlow cells, endosteum and MKs: aged: n = 10,000 measurements from 3/3/4 maps; young: n = 10,000 measurements from 3/3/5 maps. For further details about the simulation procedure and statistical analysis see Materials & Methods. See also Supplementary Figure 1 and Source Data Extended Data Fig. 6.
Extended Data Fig. 7 In young mice, transplanted HSCs lodge in at least two different functional niches, while in the aged the endosteal/arteriolar niche is functionally impaired.
a, Confocal images of whole-mount femurs from aged recipient mice transplanted with CFP+ young and YFP+ aged HSCs. Arrows (white) show donor-derived CFP+ young and YFP+ aged HSCs. b, Percentage of CFP+ young and YFP+ old HSCs in young and old recipients in proximity to arterioles, sinusoids and megakaryocytes ( < 10 µm) and in the endosteal area ( < 50 µm; n = 3 young recipient mice with, 210 CFP+ young and 30 YFP+ old HSCs from 31 areas and n = 3 old recipient mice with 80 CFP+ young and 27 YFP+ old HSCs from 30 areas). c, d, Percentage of CFP+ young and YFP+ old HSCs in the endosteal/arteriolar and sinusoidal/megakaryocytic niche in young (c) and old (d) recipients. e, f, Distribution of CFP+ young and YFP+ old HSCs to the endosteum of young (e) and old (f) recipients. g, h, Percentage of engraftment, T cells, B cells and myeloid cells from CFP+ young and YFP+ aged donor HSCs in PB of young recipient (g) (young HSCs: n = 8 mice; aged HSCs: n = 9 mice) and old recipient mice (h) (young HSCs: n = 4 mice; aged HSCs: n = 5 mice). i, Single cell RNAseq experimental layout. j-o, Histone variants (j) H2afz, (k) H3f3a and HSC regulators (l) Cxcr4, (m) Dek, (n) Gpr56 and (o) Ctnnb1 are upregulated in LR-HSCs. Values related to single cells and referred to DESeq-normalized and baselined log(FPKM). Data are plotted as average population expression ±SD (n = 28 nLR-HSCs, n = 37 LR-HSCs). Experiments in a were repeated three independent times with similar results. In b, g, h data represent mean ± s.e.m and the statistical significance was assessed by two-tailed unpaired t-tests. * p < 0.05. Scale bars, 10 μm (a). See also Source Data Extended Data Fig. 7.
Extended Data Fig. 8 Endothelial Jag2 maintains aged LR-HSCs at sinusoids.
a, Endothelial and Nes-GFPlow cells expressing Jag2 in BM of young and old Nes-GFP mice (n = 5 young, n = 3 old mice). b, Jag1 (red) in sinusoidal vessels (blue) and Nes-GFPlow cells (green) in BM from young and old Nes-GFP mice. DAPI (white). c, Overview of the in vitro co-culture experiments. d, Images showing co-culture of YFP+ HSCs (green) and endothelial cells. e, Number of YFP+ HSCs after treatment with Jag2 blocking and isotype control antibody. YFP+ HSCs samples were either assayed in co-culture with freshly prepared ECs or alone (n = 6/6/3/2). f, Representative FACS dot plots of the gating strategy for YFP+ HSCs in vitro co-culture experiments with ECs and Jag2 blocking antibody. g, BM image of a mouse treated with Jag2 blocking antibody or isotype control. Samples were stained with the same secondary antibody (red). Vessels are stained with CD31/CD144 (blue). h, Percentage of Ki-67+ and Ki-67− HSCs in distal-diaphyseal-femoral central BM areas of Jag2 blocking antibody and isotype control antibody treated mice (n = 966 and n = 109 HSCs per group from 3 areas per group; 2/3 mice/group). i, Representative 3D reconstruction of Ki-67+ (red dots) and Ki-67− (yellow dots) HSCs at sinusoids in distal-diaphyseal-femoral central BM of Jag2 blocking antibody treated mice. j-k, Percentage of Ki-67+ HSCs in proximity to sinusoids (j) and of clustered HSCs (k) in BM of mice treated with Jag2 blocking antibody and isotype control (j, 472 and 28 HSCs from n = 3 areas per group, 2/3 mice/group; k, 653 and 28 HSCs from n = 4 and n = 3 areas, 3 mice/group). Experiments in b, d, g, i were repeated three, in f 6 independent times with similar results. Data represent mean ± s.e.m. In a, j, k the statistical significance was assessed by two-tailed unpaired t-tests in e by one-way-ANOVA-test. **** p < 0.0001. Scale bars, 20 μm (b, di), 10 μm (dii), 50 µm (g, i). See also Source Data Extended Data Fig. 8.
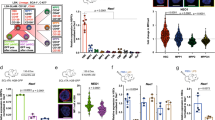
Extended Data Fig. 9 Impairment of the sinusoidal niche after 5-FU administration.
a, Experimental layout. b, Young 5-FU treated mice show Ki-67+ (green) CD150+ (red) CD41 CD48 Lin+ (white) clusters of MPPs. c, Experimental layout. d, Red blood cell count (RBC), myeloid (Gr1+, Mac1+), B cell (B220+) and T cell (Cd3+) frequencies in PB of 5-FU treated young and old mice (Young + 5FU: n = 7 mice for each panel; Aged + 5FU: n = 8 mice for each panel). e, Experimental layout of transplantations of 5-FU preconditioned, lethally irradiated (11 Gy) and control young and aged recipient mice with young or old YFP+ donor HSCs. f, Percentage of engraftment from YFP+ HSCs in PB of young and aged control and 5-FU preconditioned recipients (n = 4/6/4/6 mice). g, BV diameter per volume in stacked images in central BM (≥ 50 µm from endost) from 5-FU treated (30d) and non-treated young and old mice (135/162/151/142 BVs from n = 3 areas, two mice per group). h, Z-stacked images showing vasculature (CD31/CD144) in bones from young and old mice 30 days after 5-FU administration and non-treated controls. i, t-SNE representation of gene expression data of 1218 young and 338 aged CD31+ ECs. j, Heatmap of differentially expressed genes. 59 of 1570 genes were deemed significant using DESeq2 (FDR adjusted p-value < 0.1). Color mapping shows log-transformed, size-factor normalized and batch corrected expression. Rows were hierarchically clustered using complete linkage. k, Diffusion map representation of cell cycle states of young and aged CD31+ ECs. Cell cycle partitioning was performed based on gene expression data. l, Percentage of Ki-67+ and Ki-67− HSCs in BM images of WT young and old mice 30 days after 5-FU administration (n = 80 young HSCs and n = 156 old HSCs, 3 mice per group). m, Frequency of HSCs and LSKs among BM cells in control and 30-day-5-FU treated young and aged mice (n = 10/6/9/6 mice for HSCs, n = 4/6/4/6 mice for LSKs). Experiments in b, h were repeated three independent times with similar results. In d the statistical significance was assessed by two-way-ANOVA-test. In f, g, m data represent mean ± s.e.m and statistical significance was assessed by two-tailed unpaired t-tests. * p < 0.05, ** p < 0.01, *** p < 0.001. Scale bars, 50 μm (b); 100 μm (h). The statistical details for d are available in Source Data Extended Data Fig. 9. See also Supplementary Table 5.
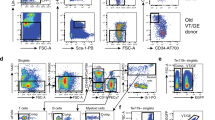
Extended Data Fig. 10 Summary cartoon scheme and gating strategies.
a, Cartoon scheme depicting alterations of the aged niches. Aged HSCs compared to young are located more distant to the endosteum, to megakaryocytes, to arterioles and to periarteriolar Nes-GFPhigh cells but not to Nes-GFPlow cells and sinusoids. With aging megakaryocytes are increased in number. Endosteal endothelial cells and endosteal/periarteriolar Nes-GFPhigh cells are dramatically reduced. The anatomy and morphology of arteries and arterioles in the aged BM is massively changed. The morphology, anatomy and numbers of central BM vessels, which are comprised mainly of sinusoids, and perisinusoidal Nes-GFPlow cells are preserved upon aging. LR-HSCs, which represent in aged mice the population of aged HSCs with the highest regenerative potential, cell polarity and active Notch signalling, were exclusively found as individual stem cells at sinusoidal niches, whereas aged nLR-HSCs are more distant to sinusoids and to other described BM niche cells and show clustering phenotype. Importantly, Jag2 plays a functional role in the maintenance of proximity and quiescence of HSCs at sinusoids. b, Representative gating strategy for Nes-GFP high and low BM cells; c, Gating strategy for megakaryocytes; d, Gating strategy for endothelial cells; e, Gating strategy for PB cells. Representative gating strategy example of B220, CD3, Mac-1 and Gr-1 staining profile of white blood cells from an aged C57Bl6 mouse (more than 100-week-old).
Supplementary information
Supplementary Information
Supplementary Figs. 1–2
Supplementary Video 1
Old LR-HSC in proximity to a sinusoid
Supplementary Video 2
Young HSC in physical association to an arteriole
Supplementary Video 3
Aged HSCs are located distant to arterioles
Supplementary Video 4
Young HSC in proximity to a sinusoid
Supplementary Video 5
Aged HSCs adjacent to a sinusoidal vessel
Supplementary Video 6
Old perisinusoidal LRC in proximity to a sinusoidal Jag2+ cell
Supplementary Tables
Supplementary Table 1: Total and endosteal numbers of young and aged HSCs. Supplementary Table 2: HSC frequencies in FACS and histological whole-mounts of long bones from old mice. Supplementary Table 3: Total numbers of young and old Ki67+ HSCs and thier distances from niche cells. Supplementary Table 4: Genes upregulated in aLR-HSCs compared with anLR-HSCs. Supplementary Table 5: Differentially expressed genes between young and aged CD31+ cells. Supplementary Table 6: All antibodies used in this study and their validation
Source data
Rights and permissions
About this article
Cite this article
Saçma, M., Pospiech, J., Bogeska, R. et al. Haematopoietic stem cells in perisinusoidal niches are protected from ageing. Nat Cell Biol 21, 1309–1320 (2019). https://doi.org/10.1038/s41556-019-0418-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0418-y
This article is cited by
-
Clonal hematopoiesis and its impact on the aging osteo-hematopoietic niche
Leukemia (2024)
-
Inflammatory signals from fatty bone marrow support DNMT3A driven clonal hematopoiesis
Nature Communications (2023)
-
Cellular plasticity of the bone marrow niche promotes hematopoietic stem cell regeneration
Nature Genetics (2023)
-
The meaning of adaptation in aging: insights from cellular senescence, epigenetic clocks and stem cell alterations
Nature Aging (2023)
-
The roles of bone remodeling in normal hematopoiesis and age-related hematological malignancies
Bone Research (2023)