Abstract
Tumours depend on altered rates of protein synthesis for growth and survival, which suggests that mechanisms controlling mRNA translation may be exploitable for therapy. Here, we show that loss of APC, which occurs almost universally in colorectal tumours, strongly enhances the dependence on the translation initiation factor eIF2B5. Depletion of eIF2B5 induces an integrated stress response and enhances translation of MYC via an internal ribosomal entry site. This perturbs cellular amino acid and nucleotide pools, strains energy resources and causes MYC-dependent apoptosis. eIF2B5 limits MYC expression and prevents apoptosis in APC-deficient murine and patient-derived organoids and in APC-deficient murine intestinal epithelia in vivo. Conversely, the high MYC levels present in APC-deficient cells induce phosphorylation of eIF2α via the kinases GCN2 and PKR. Pharmacological inhibition of GCN2 phenocopies eIF2B5 depletion and has therapeutic efficacy in tumour organoids, which demonstrates that a negative MYC–eIF2α feedback loop constitutes a targetable vulnerability of colorectal tumours.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA–sequencing data and ribosomal profiling data that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE106858. Mass spectrometry data that support the findings of this study have been deposited in figshare.com under the title of this manuscript (“A MYC–GCN2–eIF2a negative feedback loop limits protein synthesis to prevent MYC dependent apoptosis in colorectal cancer”) by W.S. (https://figshare.com/articles/A_MYC_GCN2_eIF2a_negative_feedback_loop_limits_protein_synthesis_to_prevent_MYCdependent_apoptosis_in_colorectal_cancer_Fig_5a/9878723 and https://figshare.com/articles/A_MYC_GCN2_eIF2a_negative_feedback_loop_limits_protein_synthesis_to_prevent_MYCdependent_apoptosis_in_colorectal_cancer_Fig_5c/9878717). All other data supporting the findings of this study are available from the corresponding authors upon reasonable request.
References
Truitt, M. L. et al. Differential requirements for eIF4E dose in normal development and cancer. Cell 162, 59–71 (2015).
Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010).
Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Jennings, M. D., Kershaw, C. J., Adomavicius, T. & Pavitt, G. D. Fail-safe control of translation initiation by dissociation of eIF2α phosphorylated ternary complexes. eLife 6, e24542 (2017).
Kenner, L. R. et al. eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science 364, 491–495 (2019).
Adomavicius, T. et al. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10, 2136 (2019).
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
van de Wetering, M. et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250 (2002).
Sansom, O. J. et al. Myc deletion rescues Apc deficiency in the small intestine. Nature 446, 676–679 (2007).
Dow, L. E. et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal. Cancer Cell 161, 1539–1552 (2015).
Faller, W. J. et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497–500 (2015).
Truitt, M. L. & Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 17, 332 (2017).
Truitt, M. L. & Ruggero, D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer 16, 288–304 (2016).
Barna, M. et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975 (2008).
Thoreen, C. C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Wiegering, A. et al. Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high MYC levels in colorectal cancer. Cancer Discov. 5, 768–781 (2015).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Sansom, O. J. et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 18, 1385–1390 (2004).
Willert, J. et al. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 2, 8 (2002).
Bild, A. H. et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439, 353–357 (2006).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Faux, M. C. et al. Restoration of full-length adenomatous polyposis coli (APC) protein in a colon cancer cell line enhances cell adhesion. J. Cell Sci. 117, 427–439 (2004).
Rosenbluh, J. et al. β-catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151, 1457–1473 (2012).
Jennings, M. D. & Pavitt, G. D. A new function and complexity for protein translation initiation factor eIF2B. Cell Cycle 13, 2660–2665 (2014).
Pavitt, G. D. Regulation of translation initiation factor eIF2B at the hub of the integrated stress response. Wiley Interdiscip. Rev. RNA 9, e1491 (2018).
Hart, T. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015).
Fresno, M., Jimenez, A. & Vazquez, D. Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72, 323–330 (1977).
Lobo, M. V. et al. Levels, phosphorylation status and cellular localization of translational factor eIF2 in gastrointestinal carcinomas. Histochem. J. 32, 139–150 (2000).
Crouch, D. & Safer, B. The association of eIF-2 with Met-tRNAi or eIF-2B alters the specificity of eIF-2 phosphatase. J. Biol. Chem. 259, 10363–10368 (1984).
Ingolia, N. T. et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Rubio, C. A. et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 15, 476 (2014).
Hinnebusch, A. G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 (2005).
Gardner, B. M. et al. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb. Perspect. Biol. 5, a013169 (2013).
Hetz, C., Chevet, E. & Oakes, S. A. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 17, 829–838 (2015).
Jiang, H. Y. et al. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24, 1365–1377 (2004).
Zinszner, H. et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 (1998).
Murphy, D. J. et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell 14, 447–457 (2008).
Shi, Y. et al. Therapeutic potential of targeting IRES-dependent c-myc translation in multiple myeloma cells during ER stress. Oncogene 35, 1015–1024 (2016).
Stoneley, M. et al. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 20, 1162–1169 (2000).
Didiot, M. C. et al. Identification of cardiac glycoside molecules as inhibitors of c-Myc IRES-mediated translation. J. Biomol. Screen. 18, 407–419 (2013).
Berg, J. M., Tymoczko, J. L. & Stryer, L. Biochemistry 6th edn (W. H. Freeman, 2006).
Zuber, J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–528 (2011).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Shibata, H. et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 278, 120–123 (1997).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Adhikary, S. & Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6, 635–645 (2005).
Su, L. K. et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256, 668–670 (1992).
Donnelly, N. et al. The eIF2α kinases: their structures and functions. Cell. Mol. Life Sci. 70, 3493–3511 (2013).
Haikala, H. M. et al. Pharmacological reactivation of MYC-dependent apoptosis induces susceptibility to anti-PD-1 immunotherapy. Nat. Commun. 10, 620 (2019).
Nieminen, A. I. et al. Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proc. Natl Acad. Sci. USA 110, E1839–E1848 (2013).
Castilho, B. A. et al. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta 1843, 1948–1968 (2014).
Yue, M. et al. Oncogenic MYC activates a feedforward regulatory loop promoting essential amino acid metabolism and tumorigenesis. Cell Rep. 21, 3819–3832 (2017).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Kaiser, S. et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8, R131 (2007).
Hong, Y. et al. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin. Exp. Metastasis 27, 83–90 (2010).
Eden, E. et al. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Eden, E. et al. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 3, e39 (2007).
Fellmann, C. et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol. Cell 41, 733–746 (2011).
Weber, K. et al. A multicolor panel of novel lentiviral ‘gene ontology’ (LeGO) vectors for functional gene analysis. Mol. Ther. 16, 698–706 (2008).
Herold, S. et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature 567, 545–549 (2019).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Dai, Z. et al. edgeR: a versatile tool for the analysis of shRNA-seq and CRISPR-Cas9 genetic screens. F1000Res 3, 95 (2014).
Afgan, E. et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018).
Dejure, F. R. et al. The MYC mRNA 3′-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. EMBO J. 36, 1854–1868 (2017).
Moser, A. R., Pitot, H. C. & Dove, W. F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247, 322–324 (1990).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Acknowledgements
This study was supported by grants from the following sources: Else–Kröner–Fresenius Foundation (2015_A57 to A.W.); the interdisciplinary center for clinical research of the Medical Faculty Würzburg (IZKF B-186 and B-335 to A.W.); European Research Council Grants ‘AuroMYC’ (Advanced Grant to M.E.) and ‘ColonCan’ (Starting Grant to O.J.S.; 311301); a Cancer Research UK Grand Challenge grant (A25045 to O.J.S.); Cancer Research UK core funding (A17196 and A21139 to O.J.S.); the Deutsche Forschungsgemeinschaft (DFG) (WO 2108/1-1 to E.W., FOR2314 and KFO DFG EI 222/8-1 grants to M.E., FOR2314 and KFO DFG WI 5037/2-2 to A.W.); and the Wilhelm Sander-Stiftung (to M.E.). S.W. is supported by the Comprehensive Cancer Center programme of the German Cancer Aid (Deutsche Krebshilfe). Additional personal financial support was given by S. Kratz. The technical expertise of S. Roth, B. Bauer, H. Marouf and C. Schneider is gratefully acknowledged. The invaluable support of the Histology Service, the Biological Services Unit and all the core services at the Cancer Research UK Beatson Institute is greatly appreciated (Cancer Research UK core grant C596/A17196).
Author information
Authors and Affiliations
Contributions
S.S., D.G., F.W.U., O.J.S., M.E. and A.W. conceived the project and directed experiments. S.S., D.G., F.W.U., S.D., M.P., N.M., S.B., C.F., F.C.W., C.P.A., A.B., R.J., C.F., C.S.-V., K.M., W.S., J.R.P.K., D.S., A.R. and A.W. performed the experiments. S.S., D.G., F.W.U., S.D., M.P., C.P.A., F.E., W.S., E.W., A.S., A.R., O.J.S., M.E. and A.W. analysed and interpreted the data. F.W.U., C.P.A., S.W. and F.E. performed the bioinformatics analyses. M.E.D., S.B., F.C.W., G.V., N.V., C.O., E.W., D.S., C.-T.G., A.R., O.J.S. and M.E. provided administrative, technical or material support. S.S., D.G., E.W., C.-T.G., O.J.S., M.E. and A.W. wrote the manuscript. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 An shRNA screen identifies eIF2B5 as a survival factor in APCdef cells.
(a) Diagram illustrating the generation of SW480TetOnAPC cells. (b) Immunoblot of APCdef (ethanol) and APCres cells (doxycycline for the indicated time points), representative of two independent experiments with similar results. (c) Relative mRNA expression of AXIN2 and MYC determined via qPCR treated as described in (a). Data show mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (d) Annexin V/PI FACS analysis of APCdef and APCres cells (72 h ethanol or doxycycline, respectively), representative of three independent experiments with similar results. Numbers represent percentage of cells in each quadrant. (e) Outline of shRNA screen. T0, T3 and T15 indicate time points (days) when cells were harvested for library preparation. (f) Plot showing median log2 fold change of all shRNAs included in the screen in APCres versus APCdef cells (n = 3 biologically independent experiments). (g) Plot showing median log2 fold change as described in (f) with shRNAs targeting luciferase displayed in blue. (h) Plot showing median log2 fold change as described in (f) with shRNAs targeting PSMB2 displayed in colour. (i) Plot showing median log2 fold change as described in (f) with shRNAs identified as specifically depleted in APCdef cells displayed in red. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 1.
Extended Data Fig. 2 Apoptosis induction upon EIF2B5 knockdown in APCdef cells is an on-target effect.
(a) PI cell cycle FACS analysis of shCTR-transduced or eIF2B5-depleted APCdef and APCres cells (six days ethanol or doxycycline, respectively). Data show mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (b) Immunoblot of shCTR-transduced (-) or eIF2B5-depleted APCdef and APCres cells (96 h ethanol or doxycycline, respectively), representative of two independent experiments with similar results. (c) Annexin V/PI FACS analysis of shCTR-transduced (-) or eIF2B5-depleted APCdef and APCres cells treated as described in (b). Data show mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (d) Immunoblot of control (‘empty’) and eIF2B5mut-HA overexpressing shCTR-transduced or eIF2B5-depleted APCdef and APCres cells (96 h ethanol or doxycycline, respectively), representative of two independent experiments with similar results. (e) mRNA expression of endogenous EIF2B5 of cells described in (d). Primers targeting the EIF2B5 3′UTR were used. Data show mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (f) Annexin V/PI FACS analysis of cells described in (d). Data show mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (g) Annexin V/PI FACS analysis of shCTR-transduced or eIF2B5-depleted HT29 and HCT116 cells. Data show mean ± s.d. (n = 3 biologically independent experiments); unpaired, two-tailed t-test. (h) Box plots of EIF2B5 mRNA expression in normal colorectal tissue and colorectal carcinoma of two independent datasets from Oncomine (n = number of biologically independent samples). Data sets are taken from ‘Kaiser’;56 ‘Hong’57. Points: minimum and maximum, whiskers: 10th and 90th percentile, box: 25th and 75th percentile, line: median; unpaired t-test. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 2.
Extended Data Fig. 3 eIF2B5 depletion induces an ISR.
(a) Polysome profiling of shCTR-transduced and eIF2B5-depleted APCres cells (72 h doxycycline) incubated with harringtonine for 0 s (upper) and 180 s (lower). Data (0 s harringtonine) are representative of three independent experiments with similar results, 180 s harringtonine assay was performed once. (b) 35S-methionine labelling of shCTR-transduced or eIF2B5-depleted APCdef and APCres cells (96 h ethanol or doxycycline, respectively). Incorporated radioactivity was measured by scintillation counting. Data represent mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (c) Immunoblots of cells described in (b), representative of two independent experiments with similar results. (d) Immunoblots of cells described in (b), representative of two independent experiments with similar results. p-eIF2α S51 levels, relative to total eIF2α, are shown below the immunoblot. (e) Plots documenting mean log2 fold change of total mRNA (x-axis, ‘RNA’) and ribosome-associated mRNA (y-axis, ‘Ribo’) of eIF2B5-depleted APCdef and APCres cells (96 h ethanol or doxycycline, respectively). MYC is highlighted in red (n = 3 biologically independent experiments). (f) Gene ontology (GO) analyses of ribosomal profiling data from (e). P values, adjusted for multiple testing, are shown for over-representation of GO terms among genes upregulated after EIF2B5 knockdown in APCdef and APCres cells. P values were calculated using GOrilla58,59. (g) Immunoblot of cells as described in (b), representative of two independent experiments with similar results. (h) mRNA expression of spliced XBP1, GRP78 and unspliced XBP1 of cells described in (b). Data show mean ± s.d. (n = 3 biologically independent experiments); unpaired, two-tailed t-test. (i) mRNA expression of ATF3 and GADD34 of cells described in (b). Data show mean ± s.d. (ATF3 n = 4, GADD34 n = 3 biologically independent experiments); unpaired, two-tailed t-test. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 3.
Extended Data Fig. 4 The ISR induced by eIF2B5 depletion is MYC-dependent.
(a) Immunoblot of shCTR-transduced or eIF2B5-depleted APCdef and APCres cells (96 h ethanol or doxycycline, respectively), representative of two independent experiments with similar results. For ISR induction, cells were treated with 1 µg/ml tunicamycin (3 h), DMSO was used as solvent control. (b) Immunoblot of shCTR-transduced or eIF2B5-depleted APCdef and APCres cells upon CHOP depletion (96 h ethanol or doxycycline, respectively), representative of three independent experiments with similar results. siRNA transfections were carried out using siCTR as non-targeting control or siCHOP for 72 h. (c) Annexin V/PI FACS analysis of cells treated as described in (b). Data show mean ± s.d. (n = 3 biologically independent experiments); unpaired, two-tailed t-test. (d) qPCR documenting MYC expression in shCTR-transduced or eIF2B5-depleted APCdef and APCres cells (96 h ethanol or doxycycline, respectively). Data show mean ± s.d. (n = 3 biologically independent experiments); unpaired, two-tailed t-test. (e) Immunoblot of shCTR-transduced or eIF2B5-depleted APCdef and APCres cells (72 h ethanol or doxycycline, respectively) after cycloheximide (CHX) pulse chase. Experiment was performed once. Cells were treated with 100 µg/ml CHX for the indicated time points. (f) Immunoblot of shCTR-transduced or eIF2B5-depleted APCdef and APCres cells (72 h ethanol or doxycycline, respectively), representative of three independent experiments with similar results. (g) qPCR documenting MYC expression in shCTR-transduced or eIF2B5-depleted APCdef and APCres cells treated as described in (f). Data represent mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (h) Immunoblot of shCTR-transduced or eIF2B5-depleted HT29 and HCT116 cells, representative of two independent experiments with similar results. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 4.
Extended Data Fig. 5 eIF2B5 depletion regulates IRES-mediated translation of MYC.
(a) Immunoblot of SW480 cells after DMSO (-) or cymarine ( + ) treatment (100 nM, 24 h), representative of two independent experiments with similar results. Quantification of MYC, Cyclin E and c-Fos, relative to Vinculin, is shown below the immunoblot. (b) Schematic diagram of the MYC IRES sequence. Position of used sgRNAs for deletion of the IRES as well as primers for detection by PCR and expected size of the PCR products are indicated. (c) Agarose gel electrophoresis of PCR products from MYC IRES undeleted (CTR) and deleted (sgMYC IRES) cells. The primer pair used is shown in (b). PCR was performed once as control for deletion. (d) Immunoblot of control (CTR) and MYC IRES deleted (sgMYC IRES) APCdef and APCres cells, transduced with shCTR or shEIF2B5 #3 (72 h ethanol or doxycycline, respectively), representative of two independent experiments with similar results. (e) mRNA expression of ATF3 and GADD34 in shCTR and eIF2B5-depleted APCdef and APCres cells upon MYC depletion (96 h ethanol or doxycycline, respectively). siRNA transfections were carried out using siCTR as non-targeting control or siMYC for 72 h. Data represent mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 5.
Extended Data Fig. 6 eIF2B5 depletion suppresses growth of Apc-deleted murine organoids and patient-derived organoids.
(a) Immunoblot of wild-type, VillinCreERApcfl/fl and VillinCreERApcfl/flKrasG12D/+ murine intestinal organoids. Immunoblot was performed once. (b) mRNA expression of Eif2b5 in wild-type, VillinCreERApcfl/fl and VillinCreERApcfl/flKrasG12D/+ murine intestinal organoids infected with a doxycycline-inducible shRNA targeting Eif2b5. Organoids were treated with ethanol as solvent control or 1 µg/ml doxycycline for 72 h. Data represent mean ± s.d. (n = 3 technical replicates), representative of two independent experiments with similar results. (c) Growth of murine intestinal organoids (wild-type, VillinCreERApcfl/fl and VillinCreERApcfl/flKrasG12D/+) as described in (b). Single cells were seeded and one day later treated with ethanol or 1 µg/ml doxycycline for six days. A representative picture is shown for each genotype. Scale bars = 200 µM. (d) Size of murine intestinal organoids from (c). Data represent mean ± s.e.m. (at least n = 9 biologically independent samples). Whiskers: minimum and maximum, box: 25th and 75th percentile, line: median; unpaired, two-tailed t-test. (e) Growth of shNTC-transduced or eIF2B5-depleted patient-derived CRC organoids, representative of three independent experiments with similar results. Six patient-derived organoid (T1-T5: APC-mutated; T6: APC wild-type) lines were infected with a doxycycline-inducible non-targeting control (shNTC) or an shRNA targeting EIF2B5. Organoids were treated with ethanol or 1 µg/ml doxycycline for six to seven days. Scale bars = 200 µM. (f) Quantification of viability of shNTC-transduced or eIF2B5-depleted patient-derived CRC organoids (T1, T2, T3, T4, T5) from (e). Data show mean ± s.e.m. (n = 5 biologically independent organoid lines); unpaired, two-tailed t-test. (g) Immunoblot of one patient-derived CRC organoid (T4) infected and treated as described in (e), representative of two independent experiments with similar results. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 6.
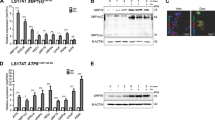
Extended Data Fig. 7 Eif2b5 heterozygosity suppresses hyperproliferation upon loss of Apc in colon.
(a) Illustration of generation of Eif2b5 knockout mice used in the study. (b) Immunohistochemical AB/PAS and lysozyme staining of wild-type and Eif2b5+/− mice in small intestinal and colon tissue, representative of three biologically independent mice of each genotype with similar results (n = 3). AB/PAS staining represents goblet cells, lysozyme staining represents Paneth cells. Scale bars = 50 µm. (c) Immunoblot of intestinal epithelial extracts from mice of the indicated genotypes. Each lane represents one separate mouse from the relevant group (left). Immunoblot was performed once. Quantification of eIF2B5 protein levels, normalised to γ-tubulin (right). Data show mean ± s.d. (n = 3 biologically independent mice); one-tailed Mann-Whitney U test. (d) Representative H&E-, eIF2B5-, p-eIF2α S51-, BrdU-, cleaved caspase 3- and MYC-stained sections of colons from mice of the indicated genotypes. Scale bars = 100 µm. (e) Graphs documenting the number of cells positive for BrdU, cleaved caspase 3, and MYC in immunostained colon sections from mice of the indicated genotypes. The number of BrdU-positive cells per half crypt (top panel), and the number of cells per full crypt staining positive for cleaved caspase 3 (middle panel) or MYC (bottom panel), were scored in 25 crypts per mouse in at least four biologically independent mice (n = 4 for BrdU staining of VillinCreERApcfl/flEif2b5+/− and VillinCreERApcfl/flKrasG12D/+, n = 5 for cleaved caspase 3 staining of VillinCreERApcfl/flEif2b5+/− and VillinCreERApcfl/flKrasG12D/+Eif2b5+/−, n = 4 for MYC staining of VillinCreERApcfl/flKrasG12D/+, n = 5 for MYC staining of wild-type, Eif2b5+/− and VillinCreERApcfl/fl, n = 6 for all other stainings and genotypes). Data show mean ± s.e.m.; one-tailed Mann-Whitney U or one-way ANOVA test. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 7.
Extended Data Fig. 8 Eif2b5 is haplo-insufficient for intestinal tumourigenesis driven by Apc loss.
(a) Immunohistochemical staining of eIF2B5 in ApcMin/+ and ApcMin/+Eif2b5+/− small intestines, representative of three biologically independent mice each with similar results. Scale bars = 100 µm. (b) Kaplan-Meier curve of ApcMin/+ (n = 14 biologically independent mice) compared with ApcMin/+Eif2b5+/− (n = 13 biologically independent mice) mice. One of the ApcMin/+ mice was censored due to a mammary tumour. P value was calculated using a Log-rank test. (c) Immunoblot of organoids established from outgrowing tumours of ApcMin/+ and ApcMin/+Eif2b5+/− mice. Each lane represents one mouse (ApcMin/+ n = 3, ApcMin/+Eif2b5+/− n = 2 biologically independent mice). Immunoblot was performed once. (d) Quantification of eIF2B5 levels, relative to β-actin, and p-eIF2α S51 levels, relative to total eIF2α, of organoids described in (c). Data show mean ± s.d. (ApcMin/+ n = 3, ApcMin/+Eif2b5+/− n = 2 biologically independent mice). (e) 35S-methionine labelling of organoids described in (c). Incorporated radioactivity was measured by scintillation counting. Data show mean of ±s.d. (n = 3 biologically independent mice); one-tailed Mann Whitney U test. (f) Quantification of polysome to sub-polysome fractions in polysome profiles from organoids described in (c). Data show mean of ±s.d. (n = 3 biologically independent mice); one-tailed Mann Whitney U test. (g) Graphs documenting the number of BrdU-positive cells per half crypt, and number of MYC-positive cells per full crypt from mice of the indicated genotypes in the small intestine. Data were scored in 25 crypts per mouse. Data show mean ± s.e.m. (n = 3 biologically independent mice); one-tailed Mann Whitney U test. (h) Representative H&E-, BrdU-, eIF2B5- and MYC-stained sections of small intestines from mice of the indicated genotypes. Mice were sampled four days post-induction. Scale bars = 100 µM. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 8.
Extended Data Fig. 9 Depletion of PKR and GCN2 kinase is compensated over time.
(a) Immunoblots of APCdef cells upon knockdown of MYC, representative of two independent experiments with similar results. siRNA transfections were carried out using siCTR as non-targeting control or siMYC for 72 h. As positive control for PKR activation, cells were treated with poly(I:C) (2 µg/ml, 4 h). (b) Crystal violet staining of shCTR-transduced, PKR- or GCN2-depleted APCdef and APCres (six days ethanol or doxycycline, respectively), representative of three independent experiments with similar results. Two independent shRNAs for both PKR and GCN2 were used. (c) Immunoblots of shCTR-transduced, PKR- or GCN2-depleted APCdef and APCres cells (96 h ethanol or doxycycline, respectively), representative of three independent experiments with similar results. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 9.
Extended Data Fig. 10 Inhibition of PKR and GCN2 reduces viability and p-eIF2α levels in patient-derived CRC organoids.
(a) Growth of patient-derived CRC organoids treated with GCN2 and PKR inhibitors, representative of three independent experiments with similar results. Seven different patient-derived organoid lines were grown for two or three days, and then treated with A-92 or C16 for 96 h at the indicated concentrations. DMSO was used as solvent control. Scale bars = 200 µM. (b) Immunoblot of three patient-derived CRC organoid lines (upper). Organoids were treated with C16 (1 µM) or A-92 (3 µM) for 6 h. DMSO (D) was used as solvent control. Quantification of p-eIF2α S51 levels, normalised to eIF2α (lower). Data show mean ± s.d. (n = 3 biologically independent organoid lines); unpaired, two-tailed t-test. (c) Annexin V/PI FACS of shCTR-transduced, PKR- or GCN2-depleted APCdef cells (96 h ethanol) after treatment with DMSO, A-92 or C16 for 48 h. Data shown mean ± s.d. (n = 3 biologically independent experiments); unpaired, two-tailed t-test. Unprocessed immunoblots are shown in Source Data Extended Data Fig. 10.
Supplementary information
Supplementary Table 1
List of gene sets corresponding to Fig. 1.
Supplementary Table 2
List of hits corresponding to Extended Data Fig. 1.
Supplementary Table 3
List of genes of ribosomal profiling experiment.
Supplementary Table 4
Genes in gene set Reactome: activation of genes by ATF4.
Supplementary Table 5
shRNA sequences.
Supplementary Table 6
siRNA sequences.
Supplementary Table 7
Primary antibodies.
Supplementary Table 8
Primer.
Supplementary Table 9
Results of panel-sequencing of patient-derived organoid lines.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 1
Uncropped blots
Source Data Fig. 2
Statistical source data
Source Data Fig. 2
Uncropped blots
Source Data Fig. 3
Statistical source data
Source Data Fig. 3
Uncropped blots
Source Data Fig. 4
Statistical source data
Source Data Fig. 4
Uncropped blots
Source Data Fig. 5
Statistical source data
Source Data Fig. 5
Uncropped blots
Source Data Fig. 6
Statistical source data
Source Data Fig. 6
Uncropped blots
Source Data Fig. 7
Statistical source data
Source Data Fig. 7
Uncropped blots
Source Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 1
Statistical source data
Source Data Extended Data Fig. 1
Uncropped blots
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 2
Uncropped blots
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 3
Uncropped blots
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 4
Uncropped blots
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 5
Uncropped blots
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 6
Uncropped blots
Source Data Extended Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 7
Uncropped blots
Source Data Extended Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 8
Uncropped blots
Source Data Extended Data Fig. 9
Uncropped blots
Source Data Extended Data Fig. 10
Statistical source data
Source Data Extended Data Fig. 10
Uncropped blots
Rights and permissions
About this article
Cite this article
Schmidt, S., Gay, D., Uthe, F.W. et al. A MYC–GCN2–eIF2α negative feedback loop limits protein synthesis to prevent MYC-dependent apoptosis in colorectal cancer. Nat Cell Biol 21, 1413–1424 (2019). https://doi.org/10.1038/s41556-019-0408-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0408-0
This article is cited by
-
Integrated whole transcriptome profiling revealed a convoluted circular RNA-based competing endogenous RNAs regulatory network in colorectal cancer
Scientific Reports (2024)
-
C16, a PKR inhibitor, suppresses cell proliferation by regulating the cell cycle via p21 in colorectal cancer
Scientific Reports (2024)
-
Amino acid metabolism in immune cells: essential regulators of the effector functions, and promising opportunities to enhance cancer immunotherapy
Journal of Hematology & Oncology (2023)
-
Metabolic reprogramming in colorectal cancer: regulatory networks and therapy
Cell & Bioscience (2023)
-
Long non-coding RNA HIF1A-As2 and MYC form a double-positive feedback loop to promote cell proliferation and metastasis in KRAS-driven non-small cell lung cancer
Cell Death & Differentiation (2023)