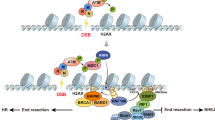
Abstract
Damage-induced long non-coding RNAs (dilncRNA) synthesized at DNA double-strand breaks (DSBs) by RNA polymerase II are necessary for DNA-damage-response (DDR) focus formation. We demonstrate that induction of DSBs results in the assembly of functional promoters that include a complete RNA polymerase II preinitiation complex, MED1 and CDK9. Absence or inactivation of these factors causes a reduction in DDR foci both in vivo and in an in vitro system that reconstitutes DDR events on nucleosomes. We also show that dilncRNAs drive molecular crowding of DDR proteins, such as 53BP1, into foci that exhibit liquid–liquid phase-separation condensate properties. We propose that the assembly of DSB-induced transcriptional promoters drives RNA synthesis, which stimulates phase separation of DDR factors in the shape of foci.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Code availability
All code used in this study is available from the corresponding author on reasonable request.
References
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
d’Adda di Fagagna, F. Living on a break: cellular senescence as a DNA-damage response. Nature Rev. Cancer 8, 512–522 (2008).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Francia, S. et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–235 (2012).
Michelini, F. et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19, 1400–1411 (2017).
Francia, S., Cabrini, M., Matti, V., Oldani, A. & d’Adda di Fagagna, F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 129, 1468–1476 (2016).
Gioia, U. et al. Pharmacological boost of DNA damage response and repair by enhanced biogenesis of DNA damage response RNAs. Sci. Rep. 9, 6460 (2019).
D’Alessandro, G. et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 9, 5376 (2018).
Lu, W. T. et al. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 9, 532 (2018).
Pryde, F. et al. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J. Cell Sci. 118, 2043–2055 (2005).
Rossiello, F. et al. DNA damage response inhibition at dysfunctional telomeres by modulation of telomeric DNA damage response RNAs. Nat. Commun. 8, 13980 (2017).
Haberle, V. & Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621–637 (2018).
Andersson, R., Sandelin, A. & Danko, C. G. A unified architecture of transcriptional regulatory elements. Trends Genet. 31, 426–433 (2015).
Kornberg, R. The molecular basis of eukaryotic transcription (Nobel Lecture). Angew. Chem. Int. Edn 46, 6956–6965 (2007).
Vernimmen, D. & Bickmore, W. A. The hierarchy of transcriptional activation: from enhancer to promoter. Trends Genet. 31, 696–708 (2015).
Lu, H. et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018).
Cho, W. K. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018).
Boehning, M. et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 25, 833–840 (2018).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855 (2018).
Berry, J., Weber, S. C., Vaidya, N., Haataja, M. & Brangwynne, C. P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl Acad. Sci. USA 112, E5237–E5245 (2015).
Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Hyman, A. A., Weber, C. A. & Julicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Kato, M. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Langdon, E. M. et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927 (2018).
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Altmeyer, M. et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 (2015).
Lee, T. I. & Young, R. A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34, 77–137 (2000).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
Sengupta, P. et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods 8, 969–975 (2011).
Jao, C. Y. & Salic, A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA 105, 15779–15784 (2008).
Galbiati, A., Beausejour, C. & d’Adda di Fagagna, F. A novel single-cell method provides direct evidence of persistent DNA damage in senescent cells and aged mammalian tissues. Aging Cell 16, 422–427 (2017).
Galbiati, A. & d’Adda di Fagagna, F. in Cellular Senescence: Methods and Protocols (ed. Demaria, M.) 11–20 (Springer, 2019).
Wang, Y. et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 163, 174–186 (2015).
Lee, K. B., Wang, D., Lippard, S. J. & Sharp, P. A. Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl Acad. Sci. USA 99, 4239–4244 (2002).
Radebaugh, C. A. et al. TATA box-binding protein (TBP) is a constituent of the polymerase I-specific transcription initiation factor TIF-IB (SL1) bound to the rRNA promoter and shows differential sensitivity to TBP-directed reagents in polymerase I, II, and III transcription factors. Mol. Cell Biol. 14, 597–605 (1994).
Bekker-Jensen, S., Lukas, C., Melander, F., Bartek, J. & Lukas, J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170, 201–211 (2005).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
Liang, L., Wang, X., Xing, D., Chen, T. & Chen, W. R. Noninvasive determination of cell nucleoplasmic viscosity by fluorescence correlation spectroscopy. J. Biomed. Opt. 14, 024013 (2009).
Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA 112, 7189–7194 (2015).
Nair, S. J. et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 26, 193–203 (2019).
Kroschwald, S., Maharana, S. & Simon, A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters 3, e201702000010 (2017).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Onuki, A. Phase Transition Dynamics (Cambridge Univ. Press, 2002).
Voorhees, P. W. Ostwald ripening of two-phase mixtures. Annu. Rev. Mater. Sci. 22, 197–215 (1992).
Zwicker, D., Decker, M., Jaensch, S., Hyman, A. A. & Julicher, F. Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc. Natl Acad. Sci. USA 111, E2636–E2645 (2014).
Berry, J., Brangwynne, C. P. & Haataja, M. Physical principles of intracellular organization via active and passive phase transitions. Rep. Prog. Phys. 81, 046601 (2018).
Caragine, C. M., Haley, S. C. & Zidovska, A. Surface fluctuations and coalescence of nucleolar droplets in the human cell nucleus. Phys. Rev. Lett. 121, 148101 (2018).
Jeng, U.-S., Esibov, L., Crow, L. & Steyerl, A. Viscosity effect on capillary waves at liquid interfaces. J. Phys. Condens. Matter 10, 4955 (1998).
Gang, H., Krall, A. H. & Weitz, D. A. Shape fluctuations of interacting fluid droplets. Phys. Rev. Lett. 73, 3435–3438 (1994).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Dupre, A. et al. A forward chemical genetic screen reveals an inhibitor of the Mre11–Rad50–Nbs1 complex. Nat. Chem. Biol. 4, 119–125 (2008).
Gunn, A. & Stark, J. M. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol. Biol. 920, 379–391 (2012).
Difilippantonio, S. et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456, 529–533 (2008).
Michelini, F. et al. From “cellular” RNA to “smart” RNA: multiple roles of RNA in genome stability and beyond. Chem. Rev. 118, 4365–4403 (2018).
Chipumuro, E. et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159, 1126–1139 (2014).
Christensen, C. L. et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 26, 909–922 (2014).
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006).
Dimitrova, N., Chen, Y.-C. M., Spector, D. L. & de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008).
Aymard, F. et al. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 24, 353–361 (2017).
Sun, D., Wu, R., Zheng, J., Li, P. & Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415 (2018).
Bouchard, J. J. et al. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol. Cell 72, 19–36 (2018).
Rulten, S. L. et al. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 42, 307–314 (2014).
Krishnakumar, R. & Kraus, W. L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 (2010).
Kilic, S. et al. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 38, e101379 (2019).
Shin, Y. et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175, 1481–1491 (2018).
Lemaitre, C. et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 28, 2450–2463 (2014).
Berkovich, E., Monnat, R. J. Jr & Kastan, M. B. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9, 683–690 (2007).
Nojima, T., Gomes, T., Carmo-Fonseca, M. & Proudfoot, N. J. Mammalian NET-seq analysis defines nascent RNA profiles and associated RNA processing genome-wide. Nat. Protoc. 11, 413–428 (2016).
Cawthon, R. M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002).
M. F. Carey, C. L. Peterson & S. T. Smale, Dignam and Roeder nuclear extract preparation. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot5330 (2009).
Zierhut, C., Jenness, C., Kimura, H. & Funabiki, H. Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol. 21, 617–625 (2014).
Alberti, S. et al. A user’s guide for phase separation assays with purified proteins. J. Mol. Biol. 430, 4806–4820 (2018).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Cerbino, R. & Trappe, V. Differential dynamic microscopy: probing wave vector dependent dynamics with a microscope. Phys. Rev. Lett. 100, 188102 (2008).
Giavazzi, F., Brogioli, D., Trappe, V., Bellini, T. & Cerbino, R. Scattering information obtained by optical microscopy: differential dynamic microscopy and beyond. Phys. Rev. E 80, 031403 (2009).
Wilkins, D. K. et al. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 38, 16424–16431 (1999).
Cseresnyes, Z., Schwarz, U. & Green, C. M. Analysis of replication factories in human cells by super-resolution light microscopy. BMC Cell Biol. 10, 88 (2009).
Huang, F. et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat. Methods 10, 653–658 (2013).
Veatch, S. L. et al. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE 7, e31457 (2012).
Acknowledgements
We thank N. G. Walter and S. Pitchiaya (Single Molecule Analysis Group and Center for RNA Biomedicine, Department of Chemistry, University of Michigan) for the generation of crucial preliminary data; all of the F.d’A.d.F. group members for support and discussions; E. Soutoglou (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France) and J. Lukas (Novo Nordisk Foundation Center for Protein Research, Copenhagen, Denmark) for reagents; C. Zeirhut (Laboratory of Chromosome and Cell Biology, The Rockefeller University, New York, USA) for reagents and discussions; F. Iannelli for bioinformatic analysis of repair products; S. Pasqualato (Biochemistry and Structural Biology Unit at the European Institute of Oncology IRCCS, Milan, Italy) for the purification of DNA fragments. F.P. was supported by a Marie Curie international mobility fellowship part of Structured International Post Doc Program; F.G. and R.C. were supported by the Regione Lombardia and Cariplo foundation (Project-2016-0998); F.G. was supported by the Associazione Italiana per Ricerca sul Cancro, (MFAG 2018-22083); F.d’A.d.F. was supported by AIRC (application numbers 12971 and 21091), the Cariplo Foundation (grant number 2014-0812), the Fondazione Telethon (GGP17111), PRIN 2010–2011 and 2015, the Italian Ministry of Education Universities and Research EPIGEN Project, InterOmics Project and AMANDA project Accordo Quadro Regione Lombardia-CNR, a European Research Council advanced grant (322726), AriSLA (project ‘DDRNA and ALS’) and AIRC Special Program 5 per mille metastases (Project-21091).
Author information
Authors and Affiliations
Contributions
F.G. conceived and performed all of the LLPS analyses of 53BP1 foci. Y.Y. performed all of the STORM experiments. U.G., together with M.G., performed timelapse experiments of 53BP1 foci treated with NH4OAc and 1,6-hexanediol, and performed comet assays and the EJ5 repair assays. V.V. conceived and performed strand-specific RT–qPCR and qPCR analysis of ChIP experiments and dilncRNA detection both in cells and in vitro. A.G. conceived and performed DIPLA analysis. S.B. performed all of the microinjections in cells and all of the 53BP1 droplets detection experiments in vitro. M.G. performed all of the timelapse experiments assisted by U.G. F.P. performed all of the FRAP experiments and confocal analysis of 53BP1 focus formation. A.O. performed all of the quantifications of confocal images. A.F. supervised F.P. for the in vitro system and nucleosome preparation and edited the manuscript. R.C. advised F.G. and edited the manuscript. D.P. supervised S.B., M.G. and A.O. and advised them on all of the imaging experiments. E.R. supervised Y.Y. and edited the manuscript. F.P. designed and performed all of the remaining experiments and wrote the manuscript. F.d’A.d.F. conceived the study and, together with F.P., assembled and revised the manuscript. All of the authors commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
F.d’A.d.F. is listed as an inventor on the patent application: PCT/EP2013/059753.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 RNAP II and PIC are recruited to DSBs as detected by ChIP in vivo and in vitro.
(a) Bar plot shows enrichment relative to the input of γH2AX, POLR2A, POLR1A, POLR3A, PIC components, MED1 and CDK9 as detected by ChIP at actin promoter. N=1. (b) Bar plot shows enrichment relative to the input of γH2AX, POLR2A, POLR1A, POLR3A, PIC components, MED1 and CDK9 as detected by ChIP at 100 bp, 2000bp, 3000bp and 10000bp distances upstream from the DSB induced by I-PpoI at an endogenous site within the DAB1 gene. N=3 (100bp, 2000bp), N=2 (3000bp) and N=1 (10000bp) independent experiments. (c) Schematic representing 100bp DNA oligonucleotide with both ends biotinylated and bound to the beads and thus not available, and 100bp DNA oligonucleotide with only one end biotinylated and one free end. (d) Analysis by immunoblotting with the indicated antibodies of pull-down experiments with biotinylated 100bp DNA oligonucleotides bound to streptavidin beads either at both ends (lane 1: blocked ends) or one end only (lane 2: free ends) incubated with HeLa nuclear extract. One tenth of the beads was used to purify DNA and it was run on a gel to demonstrate equal amounts, N=3 independent experiments. (e) Analysis by immunoblotting with the indicated antibodies of pull-down carried out in Hela nuclear extract with biotinylated 50bp DNA oligonucleotides bound to streptavidin beads either at both ends (lane 1: blocked ends), on only one end (lane 2: blunt ends), and exposing different DNA ends (lane 3: 3’ protruding, lane 4: 5’ protruding; both 10nt protruding). One tenth of the beads was used to purify DNA and it was run on a gel to demonstrate equal amounts of DNA, N=2 independent experiments. Statistics source data are provided in Supplementary Table 2.
Supplementary Figure 2 RNAP II and PIC components localize to DSB as detected by super-resolution imaging and DI-PLA.
(a-f) Representative super-resolution images of indicated proteins in U2OS cells mock treated (a-c) or treated with NCS (d-f) for 30 min and labelled with EU (15 min pulse). (g) and (h) Cross-PC between POLR2A-pS5/TBP/CDK and γH2AX was plotted comparing to that with randomized nuclei (RND-ctrl). The box indicates mean +/- SD. n=189, 130, and 117 nuclei were collected from 4, 3, and 3 biologically independent experiments for cross-PC analyses between POLR2A-pS5/TBP/CDK7 and γH2AX, respectively, in (g) and n=184, 109, and 102 nuclei were collected from 4, 3, and 3 independent experimentsfor cross-PC analyses between POLR2A-pS5/TBP/CDK7 and γH2AX, respectively, in (h). P-values of the two-sample unpaired t-test were shown in the figure accordingly. (i-l) Co-Localization Occurrence between indicated protein pairs plotted as the function of the image density of γH2AX. The Co-Localization Occurrence was calculated as Corr(A, B)×ρ (A)×ρ (B) where Corr(A, B) is the Cross-PC between A and B in one image, and ρ(A) and ρ(B) are the image density of A and B, respectively. From i to l, n=180, 129, 104, and 391 nuclei were collected from 4, 3, 3, and 10 independent experimentsfor mock data, respectively, and n=175, 90, 98, and 320 nuclei were collected from 4, 3, 3, and 10 independent experiments for NC+S data. Note that the number of nuclei for co-localization analyses are slightly less than that submitted for Cross-PC analyses due to the failure in Auto-PC fit for some nuclei. (m) DI-PLA between CyclinA and biotin, in not irradiated Hela cells (No IR) or exposed to ionizing radiations (IR 2Gy) and fixed 30 minutes after treatment. DNA stained by DAPI. Quantifications show SEM of >75 nuclei from N=3 independent experiments. Statistics source data are provided in Supplementary Table 2.
Supplementary Figure 3 MRN, TBP and TFIIB knock-downs prevent dilncRNA synthesis without affecting other protein’s level.
(a) Immunoblotting analysis of proteins’ levels in response to siRNA treatment. Specific antibodies for target proteins are indicated. Experiment was repeated twice with similar results. (b) Immunoblotting analysis of proteins’ levels in response to siRNA treatment. Specific antibodies for target proteins are indicated. (right) dilncRNA induction upon DSB formation, at DAB1 endogenous site in HeLa cells treated with indicated siRNA, quantified by strand-specific RT-qPCR, is shown. Data shown are one representative of N=3 biologically independent experiments except for siTFIIB N=1. Statistics source data are provided in Supplementary Table 2.
Supplementary Figure 4 Inactivation by RNA interference or small molecules reduces DDR signalling in cultured cells.
(a-b) Representative figures of pATM and MDC1 staining in siLuc, siTBP and siTFIIB treated U2OS cells, irradiated with 1 Gy. Quantifications show SEM of: for pATM siLUC n=121, siTBP n=112, siTFIIB n=130; for MDC1 siLUC n=111, siTBP n=129, siTFIIB n=122. Each from 2 biologically independent experiments. γH2AX quantification included in Fig. 4a. (c) Vectors expressing knockdown-resistant TBP or TFIIB fused to GFP, or GFP alone, were transfected in U2OS cells treated with siLuc, siTBP or siTFIIB. Quantifications of 53BP1 and γH2AX foci performed on GFP positive cells are shown, error bars represent the SEM of (from top to bottom) 64, 30, 32, 53, 24, 27, 38 nuclei from 2 biologically independent experiments except for siLUC+TBPGFP and siLUC+TFIIBGFP, performed once. (d) Calculation of fraction of γH2AX foci associated with de novo transcription (EU pulse) in cells treated with siRNA against Luciferase (CTRL), TBP and TFIIB. The level of association is normalized on CTRL. The bars represent the SD and n=300, 295, and 297 nuclei from 4 independent experiments. P-values of two-sample unpaired t-test are shown in the figure accordingly. (e) Pearson correlation between EU signal in nuclei (global) and at DSB (local) upon siRNA treatment against Luciferase (CTRL), TBP and TFIIB. Canonical transcription dependent on PIC and transcription at DSB decrease proportionally. n=390, 385, and 386 nuclei from 4 independent experiments. (f) Representative figures of irradiated U2OS cells (1 Gy) and pATM staining in response to THZ1, DRB or ATM inhibitor (KU-60019) treatments are shown. Quantifications show SEM of DMSO n=114, THZ1 n=112, DRB n=114, ATMi n=110 nuclei scored from 2 biologically independent experiments γH2AX quantification included in Fig. 4b. In panels (a), (b), (c) and (f) P values were calculated by one-way ANOVA and significance are represented as ** P<0.01, **** P<0.0001. Statistics source data are provided in Supplementary Table 2.
Supplementary Figure 5 53BP1 phase separation properties and analysis.
(a) Schematic showing structured domains contained in 53BP1 proteins, together with Low Complexity Regions (adapted from S.M.A.R.T. EMBL software database https://smart.embl.de). (b) Graph plotting intrinsic disordered regions (y-axis) of human (left) and mouse (right) 53BP1 amino acid sequence (x-axis) analysed by PONDR VSL2. Scores higher than 0.5 indicate predicted disordered regions. (c) Fourier analysis of droplet shape fluctuations. (i) Differential image obtained by subtracting to an instantaneous snapshot of a large droplet its time-averaged profile. Positive regions (outlined in yellow) correspond to portions of the droplet that are outside the average contour, while negative regions (blue) corresponds to local inflections of the droplet membrane. For both axes, a unit corresponds to 1 μm. (ii) Spatial-temporal map of droplet shape fluctuation, obtained by considering, as a function of time, the value corresponding to the differential image (like the one shown in panel (a)) along a curvilinear coordinate s following the average droplet contour. Colormap is as in panel (a). (iii) Structure functions D(q,Δt) obtained from the spatio-temporal map shown in (b), and quantifying the temporal relaxation of Fourier modes of the droplet shape fluctuations. Five excitations modes are shown, corresponding to wave-vectors q=2πn/L, where L=11 μm is the length of the average droplet contour and n=1-5. Continuous lines are exponential fit to the data. (d) Visualization of turbidity associated with phase separation in in vitro reactions upon incubation at 4 °C for 48 h. Graph on the right shows quantifications of turbidity of the solutions as measured by absorbance at 350 nm wavelength. N=3 independent samples. Statistics source data are provided in Supplementary Table 2.
Supplementary Figure 6 DBSCAN clustering analysis.
(a) schematic illustration of estimation of the fraction of protein A associated with protein B. The STORM data of the two proteins are submitted for DBSCAN clustering. Each cluster of protein A is scored as a paired one If its Nearest-Neighbouring Distance (NND) to any cluster of protein B is smaller than the average radius of protein A clusters. The average radius of clusters of protein A is estimated with the Pair-Correlation function (see section in Methods). Meanwhile, clusters of both protein A and protein B are randomly traslocted and orientated, and each cluster of protein A is scored whether it is paired to protein B at such randomization level. Fraction of protein A associated with protein B is then calculated as indicated. (b) Exambples of DBSCAN clustering and cluster randomization. (c) Fraction of γH2AX with either MCM (black, negative control) or EU (red). Box denotes mean +/- SD (n = 40 and 162 nuclei collected from 2 and 4 experimental replicates for MCM and EU, respectively. The p-value of the two-sample unpaired t-test is 2E-106).
Supplementary Figure 7 MRN and PIC complexes interactions.
Unprocessed blots for Fig. 3c.
Supplementary information
Supplementary Information
Supplementary Figures 1–7, titles and legends for Supplementary Tables 1–3 and Supplementary Videos 1–6.
Supplementary Table 1
Primers and oligos.
Supplementary Table 2
Statistics source data.
Supplementary Table 3
STORM antibodies.
Supplementary Video 1
53BP1 foci are sensitive to agents that target liquid compartments.
Supplementary Video 2
53BP1 foci nucleate, grow and coarse in time.
Supplementary Video 3
53BP1 foci coalescence as liquid droplets.
Supplementary Video 4
53BP1 liquid properties are dependent on transcription.
Supplementary Video 5
Extensive treatment with THZ1 is not lethal for cells.
Supplementary Video 6
53BP1 foci rely on RNA multi-interaction properties.
Rights and permissions
About this article
Cite this article
Pessina, F., Giavazzi, F., Yin, Y. et al. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol 21, 1286–1299 (2019). https://doi.org/10.1038/s41556-019-0392-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0392-4
This article is cited by
-
Phase separation-mediated biomolecular condensates and their relationship to tumor
Cell Communication and Signaling (2024)
-
AIRE relies on Z-DNA to flag gene targets for thymic T cell tolerization
Nature (2024)
-
Liquid–liquid phase separation in Alzheimer’s disease
Journal of Molecular Medicine (2024)
-
Extracellular vesicle-packaged circBIRC6 from cancer-associated fibroblasts induce platinum resistance via SUMOylation modulation in pancreatic cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
Integrative modeling of lncRNA-chromatin interaction maps reveals diverse mechanisms of nuclear retention
BMC Genomics (2023)