Abstract
Chromosome translocation is a major cause of the onset and progression of diverse types of cancers. However, the mechanisms underlying this process remain poorly understood. Here, we identified a non-homologous end-joining protein, IFFO1, which structurally forms a heterotetramer with XRCC4. IFFO1 is recruited to the sites of DNA damage by XRCC4 and promotes the repair of DNA double-strand breaks in a parallel pathway with XLF. Interestingly, IFFO1 interacts with lamin A/C, forming an interior nucleoskeleton. Inactivating IFFO1 or its interaction with XRCC4 or lamin A/C leads to increases in both the mobility of broken ends and the frequency of chromosome translocation. Importantly, the destruction of this nucleoskeleton accounts for the elevated frequency of chromosome translocation in many types of cancer cells. Our results reveal that the lamin A/C–IFFO1-constituted nucleoskeleton prevents chromosome translocation by immobilizing broken DNA ends during tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The structure of the XRCC4–IFFO1 complex was deposited into the PDB under the accession code 6ABO. Mass spectrometry data for IFFO1-binding peptides have been deposited into FigShare with the Digital Object Identifier https://doi.org/10.6084/m9.figshare.9413870 . The mass spectrometry data for peptides associated with XRCC4, XLF and PAXX that were reanalysed in the current study are available in a previous report22. The human cancer data were derived from the TCGA Research Network (http://cancergenome.nih.gov/). The dataset derived from this resource that supports the findings of this study is available in Supplementary Table 5. The source data for Figs. 3b,d,g,h, 4a–c,e–i, 6b–d,g,j,l,n–p, 7b–g, 8b,c and Supplementary Figs. 3d,g, 4e, 5c, 6a,b,d–f, 7a–i, 8a–f have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Mitelman, F., Johansson, B. & Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 7, 233–245 (2007).
Roukos, V., Burman, B. & Misteli, T. The cellular etiology of chromosome translocations. Curr. Opin. Cell Biol. 25, 357–364 (2013).
Lieber, M. R. Mechanisms of human lymphoid chromosomal translocations. Nat. Rev. Cancer 16, 387–398 (2016).
Roukos, V. & Misteli, T. The biogenesis of chromosome translocations. Nat. Cell Biol. 16, 293–300 (2014).
Zhang, Y. et al. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv. Immunol. 106, 93–133 (2010).
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Ferguson, D. O. et al. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl Acad. Sci. USA 97, 6630–6633 (2000).
Difilippantonio, M. J. et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature 404, 510–514 (2000).
Karanjawala, Z. E., Grawunder, U., Hsieh, C. L. & Lieber, M. R. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 9, 1501–1504 (1999).
Gao, Y. et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404, 897–900 (2000).
Yan, C. T. et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc. Natl Acad. Sci. USA 103, 7378–7383 (2006).
Boboila, C. et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl Acad. Sci. USA 107, 3034–3039 (2010).
Callen, E. et al. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol. Cell 34, 285–297 (2009).
Wang, J. H. et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature 460, 231–236 (2009).
Ramiro, A. R. et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature 440, 105–109 (2006).
Yan, C. T. et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482 (2007).
Franco, S. et al. DNA-PKcs and Artemis function in the end-joining phase of immunoglobulin heavy chain class switch recombination. J. Exp. Med. 205, 557–564 (2008).
Weinstock, D. M., Brunet, E. & Jasin, M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat. Cell Biol. 9, 978–981 (2007).
Simsek, D. & Jasin, M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat. Struct Mol. Biol. 17, 410–416 (2010).
Roukos, V. et al. Spatial dynamics of chromosome translocations in living cells. Science 341, 660–664 (2013).
Soutoglou, E. et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9, 675–682 (2007).
Xing, M. et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 6, 6233 (2015).
Rual, J. F. et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178 (2005).
Dittmer, T. A. et al. Systematic identification of pathological lamin A interactors. Mol. Biol. Cell 25, 1493–1510 (2014).
Ropars, V. et al. Structural characterization of filaments formed by human Xrcc4–Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc. Natl Acad. Sci. USA 108, 12663–12668 (2011).
Junop, M. S. et al. Crystal structure of the Xrcc4 DNA repair protein and implications for end joining. EMBO J. 19, 5962–5970 (2000).
Hammel, M. et al. XRCC4 protein interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double strand break repair. J. Biol. Chem. 286, 32638–32650 (2011).
Wu, P. Y. et al. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol. Cell. Biol. 29, 3163–3172 (2009).
Sibanda, B. L. et al. Crystal structure of an Xrcc4–DNA ligase IV complex. Nat. Struct. Biol. 8, 1015–1019 (2001).
Gonzalo, S. & Kreienkamp, R. DNA repair defects and genome instability in Hutchinson–Gilford Progeria Syndrome. Curr. Opin. Cell Biol. 34, 75–83 (2015).
Kochin, V. et al. Interphase phosphorylation of lamin A. J. Cell Sci. 127, 2683–2696 (2014).
Gonzalez-Suarez, I. et al. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 28, 2414–2427 (2009).
Redwood, A. B. et al. A dual role for A-type lamins in DNA double-strand break repair. Cell Cycle 10, 2549–2560 (2011).
Gibbs-Seymour, I., Markiewicz, E., Bekker-Jensen, S., Mailand, N. & Hutchison, C. J. Lamin A/C-dependent interaction with 53BP1 promotes cellular responses to DNA damage. Aging Cell 14, 162–169 (2015).
Kind, J. et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153, 178–192 (2013).
Mahen, R. et al. A-type lamins maintain the positional stability of DNA damage repair foci in mammalian nuclei. PLoS One 8, e61893 (2013).
Lottersberger, F., Karssemeijer, R. A., Dimitrova, N. & de Lange, T. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell 163, 880–893 (2015).
Soni, A. et al. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 42, 6380–6392 (2014).
Campan, M. et al. Genome-scale screen for DNA methylation-based detection markers for ovarian cancer. PLoS One 6, e28141 (2011).
Yang, L. et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153, 919–929 (2013).
Stransky, N., Cerami, E., Schalm, S., Kim, J. L. & Lengauer, C. The landscape of kinase fusions in cancer. Nat. Comm. 5, 4846 (2014).
Gao, Q. et al. Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 23, 227–238 (2018).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015).
Schrank, B. R. et al. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66 (2018).
Caridi, C. P. et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559, 54–60 (2018).
Mahaney, B. L., Hammel, M., Meek, K., Tainer, J. A. & Lees-Miller, S. P. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem. Cell Biol. 91, 31–41 (2013).
Lisby, M., Antunez de Mayolo, A., Mortensen, U. H. & Rothstein, R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2, 479–483 (2003).
Lobachev, K., Vitriol, E., Stemple, J., Resnick, M. A. & Bloom, K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 14, 2107–2112 (2004).
Kaye, J. A. et al. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 14, 2096–2106 (2004).
Hutchison, C. J. Do lamins influence disease progression in cancer? Adv. Exp. Med. Biol. 773, 593–604 (2014).
Guo, R., Xu, D. & Wang, W. Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods 48, 72–79 (2009).
Wang, Q. S. et al. The macromolecular crystallography beamline of SSRF. Nucl. Sci. Tech. 26, 12–17 (2015).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Pettersen, E. F. et al. UCSF chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Iiizumi, S. et al. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques 41, 311–316 (2006).
Xu, D. et al. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev. 22, 2843–2855 (2008).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Jaqaman, K. et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008).
Shao, S. et al. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 44, e86 (2016).
Acknowledgements
We thank W. Wang for his advice and revisions to the manuscript, R. Xi for his advice on the analysis of genomic rearrangements, T. Misteli for providing NIH2/4 cells and D. C. van Gent for the pDVG94 plasmid. We thank the staff at the BL17U1 beamline of the Shanghai Synchrotron Radiation Facility for assistance during data collection, the mass spectrometry facility of the National Center for Protein Sciences at Peking University for assistance with the identification of the proteins and the Core Facility of Life Sciences at Peking University for assistance with imaging. This work was supported in part by the National Natural Science Foundation of China (grant numbers 31661143040, 81672773 and 31870807) and the Qidong-SLS Innovation Fund.
Author information
Authors and Affiliations
Contributions
W.L., R.G., Y.L. and D.X. designed and directed the project. W.L. and X.B. performed all experiments except the following. L.L., J.Li and Y.L. designed and performed crystal structure experiments. Y.Z. performed some of the DT40 experiments. J.Liu and H.Z. performed some of the imaging experiments. M.D. and Y.S. contributed to the imaging experiments. Q.W. and J.J. performed some of the mass spectrometry experiments. F.-Y.S., M.H. and G.G. analysed the mass spectrometry data. W.L., J.Li., Y.L. and D.X. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Sequence alignment of IFFO1 homologues.
IFFO1 orthologues were identified using the BLASTP algorithm, which searches the NR database maintained by NCBI. Human (hu, Homo sapiens AAI10388.1), mouse (mu, Mus musculus Q8BXL9.1), chicken (ch, Gallus gallus XP_015149162.1), frog (xp, Xenopuslaevis XP_002941733.1) and zebrafish (fish, Danio rerio XP_690165.5) orthologues are shown. The identical and conserved amino acids are highlighted with red letters on a yellow background and dark blue letters on a blue background, respectively, weakly similar amino acids are highlighted on a green background, and green letters indicate blocks of similar amino acids. The red boxes indicate the 4 coiled-coil motifs of the intermediate filament family protein. The blue box (NLS) indicates a potential nuclear localization signal. The residues mutated in m1-5 and N73D/R85H (see below in Supplementary Fig. 2l, m) are marked with asterisks.
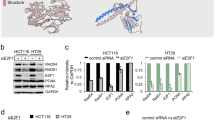
Supplementary Figure 2 Mapping the interacting regions of IFFO1 with XRCC4 and Lamin A/C.
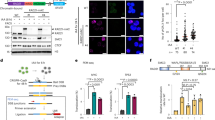
a, Immunoblotting showing that the Superose 6 fractionation profile of IFFO1 overlaps with those of XRCC4. The fractions corresponding to molecular weight standards are indicated at the top. Inputs of some proteins were cut from the right side of the same gel. b, d Schematic representations of the different IFFO1 (b) and XRCC4 (d) deletion mutants (left) and their ability to coimmunoprecipitate with XRCC4 and IFFO1 (right), respectively. FL, full length. (c, e) Immunoprecipitation to assess whether the various deletion mutants of IFFO1 (c) and XRCC4 (e) co-purified with XRCC4 and IFFO1, respectively. NA, not available. f, Immunoblot showing the IPs of Flag-tagged Lamin A and Lamin C. g, i, Schematic representations of the different Lamin A (g) and IFFO1 (i) deletion mutants (left) and their ability to interact with IFFO1 and Lamin A (right), respectively. FL, full length. h, j, Yeast two-hybrid to assess whether the various mutants of Lamin A (h) and IFFO1 (j) interacted with IFFO1 and Lamin A, respectively. k, Schematic representations of mutations of IFFO1 in cancers. Vertical axis presents the mutation frequency of the indicated residues. l, GST-pulldown to assess whether the recurrent mutations of IFFO1 affect its interaction with Lamin A. The 1A motif of IFFO1 and 2B motif of Lamin A were fused with GST-tag and MBP-tag, respectively, and expressed in E. coli for pulldown assays. The pulldown proteins were analysed by Coomassie Blue-stained SDS–PAGE gels. m, Immunoblotting to show the immunoprecipitates of FLAG-IFFO1 wild-type and mutants using HEK293 cells. All immunoblots, gels and yeast two-hybrid are representative of three independent experiments; unprocessed scans of immunoblots are shown in Supplementary Fig. 9.
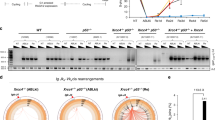
Supplementary Figure 3 Generation of IFFO1 and LMNA knockout DT40 cells.
a, Schematic representation of the chicken IFFO1 and targeted genomic DNA. The regions containing exon1-7 (marked by red) of the gene are replaced by blasticidine and histidinol resistant genes. The two regions between two pairs of dotted lines were used as arms of the knockout constructs. Arrows indicated targeted location of primers for genomic PCR. b, Genome-PCR analysis to show that IFFO1 gene is undetectable in the respective knockout DT40 cells. XLF was included as a positive control. c, Immunoblotting to show that IFFO1 protein is absent in the knockout cells. d, Sensitivity assay of the IFFO1-/- DT40 cells to various DSB-induced agents. Bars represent mean ± SD; n = 3 independent experiments. e, Schematic representation of the chicken LMNA and targeted genomic DNA. The regions containing exon2-7 (marked by red) of the gene are replaced by blasticidine, histidinol and puromycin resistant genes. The two regions between two pairs of dotted lines were used as arms of the knockout constructs. Arrows indicated targeted locations of primers for genomic PCR. f, Genome-PCR analysis to show that LMNA gene is undetectable in the respective knockout DT40 cells. XRCC4 was included as a positive control. g, Sensitivity assay of the LMNA-/-/- DT40 cells to various DSB-induced agents. Bars represent mean ± SD; n = 3 independent experiments. All immunoblots and gels are representative of three independent experiments; unprocessed scans of immunoblots are shown in Supplementary Fig. 9. Statistical source data including precise P values are shown in Supplementary Table 5.
Supplementary Figure 4 Ectopically expressing XLF or PAXX could not suppress the mobility of the broken ends in the IFFO1-/- cells.
a, Immunoblotting show the expression level of IFFO1 in the IFFO1-/- cells transfected with vary versions of IFFO1. b, c, Immunoblottings show the knockdown efficiency of XRCC4 (b) and Lamin A/C (c). Two shRNAs were used for XRCC4 (shX4_1/2) and Lamin A/C (shLM_1/2), respectively. d, Immunoblotting show the expression level of XLF and PAXX in the IFFO1-/- cells. e, MSD of the mCherry–53BP1TD foci in the IFFO1-/- cells transfected with XLF or PAXX. Means with SEMs of every time point are showed. The numbers of traces pooled from two independent experiments are indicated. All immunoblots are representative of three independent experiments; unprocessed scans of immunoblots are shown in Supplementary Fig. 9. Statistical source data including precise P values are shown in Supplementary Table 5.
Supplementary Figure 5 Lig3 promotes chromosome translocation in the IFFO1-/- cells.
a, Schematic of the pDVG94 plasmid used to examine the efficiency of aNHEJ. This figure was adapted from Verkaik NS, et.al (Eur J Immunol. 2002). b, The indicated cell lines were treated with or without 10 μM of the DNA-PKcs inhibitor, NU7441, for 4 hr, and then transfected with EcoRV- and AfeI-linearized pDVG94. Cells were allowed 24 hr to repair the linearized template, then plasmids were extracted, and the region spanning the cut site was amplified by PCR, followed by digestion with BstXI. The restriction enzyme products were then analysed by PAGE gel. c, Chromosome translocation of the IFFO1-/- cells transfected with Lig3 or Lig4 shRNA. Bars represent means with SEM of 4 independent experiments. Two-tailed t-tests were used for the analysis. **, P<0.01; ns, P>0.05. Western blotting at right panel showed the protein levels. All immunoblots and gels are representative of three independent experiments; unprocessed scans of immunoblots are shown in Supplementary Fig. 9.
Supplementary Figure 6 IFFO1 expression is downregulated by its promoter methylation in multiple tumour types.
a, The correlation between the IFFO1 expression and its methylation at promoter. Correlation Coefficient (R) and P Value (PR) were calculated using Pearson Correlation; n = 16 cancer types. b, Relative expression in tumour vs. normal tissue in multiple tumour types. Gene expression data in TCGA were accessed through The UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/). c, Heatmaps of DNA methylation at IFFO1 promoter in different cell lines. Methylation of 18 CpG sites in chr.12: 6,556,011-6,556,233 (GRCh38.p12) were determined. Red and cyan squares denote methylated and non-methylated CpG sites, respectively. White squares denote CpG sites, which were not covered by sequencing. d, mRNA of IFFO1 was measured by qPCR. GAPDH mRNA was measured as a control. Means with SD from n = 3 independent experiments were present. Two-tailed t-tests were used for the analysis. e, Correlation between IFFO1 expression and its promoter methylation in these cell lines examined. Analysis was performed using Pearson Correlation; n= 9 cell lines. f, IFFO1 mRNA level after azacytidine treatment. The EKVX cells were treated with azacytidine for 3 days before harvest. Means with SD from n = 3 independent experiments were present. Two-tailed t-tests were used for the analysis. Statistical source data including precise P values are shown in Supplementary Table 5.
Supplementary Figure 7 IFFO1 expression negatively correlates with the frequency of gene fusion in multiple cancer types.
a, The frequency of inter-chromosome translocation in multiple cancer types. The data were gained from the literature by Yang et al., 2013. OV, ovarian serous cystadenocarcinoma; GBM, glioblastoma multiforme; MM, multiple myeloma. b, Plot shows the correlation between the IFFO1 expression and the frequency of inter-chromosome translocation. Correlation Coefficient (R) and P Value (PR) were calculated using Pearson Correlation; n = 7 cancer types. c, Graph showing the average numbers of kinase fusions per sample in the multiple tumour types described by Stransky et al. 2014. d, Plot shows the correlation between the kinase fusion and the inter-chromosome translocation. Correlation Coefficient (R) and P Value (PR) were calculated using Pearson Correlation; n = 9 cancer types. e, Plots show the correlation between gene expression and the frequency of the kinase fusion in multiple tumour types, including UCEC, BLCA, LUSC, COAD, READ, LUAD, PRAD, BRCA, THCA, HNSC, LIHC, KIRP and KIRC. Correlation Coefficient (R) and P Value (PR) were calculated using Pearson Correlation; n = 13 cancer types. f, h, The frequency of kinase fusion (f) and total gene fusion (h) in multiple cancer types. The data were gained from the literature by Gao et al., 2018. g, i, Plots show the correlation between the inter-chromosome translocation with the frequency of kinase fusion (g) and total gene fusion (i). Correlation Coefficient (R) and P Value (PR) were calculated using Pearson Correlation; n = 9 cancer types. Statistical source data including precise P values are shown in Supplementary Table 5.
Supplementary Figure 8 The expression and methylation levels of IFFO1 and LMNA in multiple cancer cell lines from The Cancer Cell Line Encyclopedia (CCLE).
a-c, The expression (a) and methylation (b) levels of IFFO1 gene and their correlation (c) in CCLE. Correlation Coefficient (R) and P Value (PR) were calculated using Pearson Correlation; n = 834 cell lines (c). Sample sizes in (a, b) were indicated in the brackets. d-f, The expression (d) and methylation (e) levels of LMNA gene and their correlation (f) in CCLE. Correlation Coefficient (R) and P Value (PR) were calculated using Pearson Correlation; n = 234 cell lines (f). Sample sizes in (d, e) were indicated in the brackets. g, Plot of expressions of IFFO1 and LMNA in cancer cell lines of CCLE. Red and blue boxes indicate IFFO1-positive LMNA-negative and IFFO1-negative LMNA-positive cell lines, respectively. Most IFFO1-positive LMNA-negative cells belong hematological malignancies. All the CCLE data and analysis were accessed through the website (https://portals.broadinstitute.org/ccle). Statistical source data including precise P values are shown in Supplementary Table 5.
Supplementary Figure 9 Uncropped images from gels and blots.
Asterisks indicate the inputs.
Supplementary information
Supplementary Information
Supplementary Figures 1–9, Supplementary Note 1 and Supplementary Table titles.
Rights and permissions
About this article
Cite this article
Li, W., Bai, X., Li, J. et al. The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis. Nat Cell Biol 21, 1273–1285 (2019). https://doi.org/10.1038/s41556-019-0388-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0388-0
This article is cited by
-
LAP2α preserves genome integrity through assisting RPA deposition on damaged chromatin
Genome Biology (2022)
-
Ku70 affects the frequency of chromosome translocation in human lymphocytes after radiation and T-cell acute lymphoblastic leukemia
Radiation Oncology (2022)
-
Centrosome, microtubule and DNA damage response
Genome Instability & Disease (2022)
-
Genome-Wide Association and Expression Analysis Revealed the Candidate Variants and Molecular Underpinnings of Cold-Stress Response in Large Yellow Croaker
Marine Biotechnology (2022)
-
Rare variants in IFFO1, DTNB, NLRC3 and SLC22A10 associate with Alzheimer’s disease CSF profile of neuronal injury and inflammation
Molecular Psychiatry (2022)