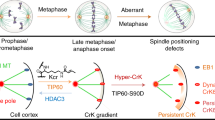
Abstract
One of the first steps in mitotic spindle assembly is the dissolution of the centrosome linker followed by centrosome separation driven by EG5, a tetrameric plus-end-directed member of the kinesin-5 family. However, even in the absence of the centrosome linker, the two centrosomes are kept together by an ill-defined microtubule-dependent mechanism. Here we show that KIFC3, a minus-end-directed kinesin-14, provides microtubule-based centrosome cohesion. KIFC3 forms a homotetramer that pulls the two centrosomes together via a specific microtubule network. At mitotic onset, KIFC3 activity becomes the main driving force of centrosome cohesion to prevent premature spindle formation after linker dissolution as it counteracts the increasing EG5-driven pushing forces. KIFC3 is eventually inactivated by NEver in mitosis-related Kinase 2 (NEK2) to enable EG5-driven bipolar spindle assembly. We further show that persistent centrosome cohesion in mitosis leads to chromosome mis-segregation. Our findings reveal a mechanism of spindle assembly that is evolutionary conserved from yeast to humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Previously published RNA-sequencing data that were reanalysed here are available under accession codes GSE11148538 and GSE6057039. Source data for Figs. 1–3 and 5–8 and Supplementary Figs. 1–8 have been provided in Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
The FIJI or ImageJ macros used in this study are available from the corresponding author on request.
References
Bornens, M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25–34 (2002).
Wang, W. J., Soni, R. K., Uryu, K. & Tsou, M. F. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739 (2011).
Bornens, M. The centrosome in cells and organisms. Science 335, 422–426 (2012).
Tsou, M. F. & Stearns, T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951 (2006).
Schmidt, K. N. et al. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199, 1083–1101 (2012).
Tanos, B. E. et al. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163–168 (2013).
Delgehyr, N., Sillibourne, J. & Bornens, M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118, 1565–1575 (2005).
Bahe, S., Stierhof, Y. D., Wilkinson, C. J., Leiss, F. & Nigg, E. A. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 171, 27–33 (2005).
Graser, S., Stierhof, Y. D. & Nigg, E. A. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 120, 4321–4331 (2007).
Fry, A. M. et al. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563–1574 (1998).
Vlijm, R. et al. STED nanoscopy of the centrosome linker reveals a CEP68-organized, periodic rootletin network anchored to a C-Nap1 ring at centrioles. Proc. Natl Acad. Sci. USA 115, E2246–E2253 (2018).
Nigg, E. A. & Stearns, T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. C ell Biol. 13, 1154–1160 (2011).
Panic, M., Hata, S., Neuner, A. & Schiebel, E. The centrosomal linker and microtubules provide dual levels of spatial coordination of centrosomes. PLoS Genet. 11, e1005243 (2015).
Mayor, T., Hacker, U., Stierhof, Y. D. & Nigg, E. A. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J. Cell Sci. 115, 3275–3284 (2002).
Mardin, B. R. et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176 (2010).
Mardin, B. R., Agircan, F. G., Lange, C. & Schiebel, E. Plk1 controls the Nek2A-PP1γ antagonism in centrosome disjunction. Curr. Biol. 21, 1145–1151 (2011).
Sawin, K. E., LeGuellec, K., Philippe, M. & Mitchison, T. J. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359, 540–543 (1992).
Nam, H. J. & van Deursen, J. M. Cyclin B2 and p53 control proper timing of centrosome separation. Nat. Cell Biol. 16, 538–549 (2014).
Kaseda, K., McAinsh, A. D. & Cross, R. A. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol. Open 1, 12–18 (2012).
Blangy, A. et al. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83, 1159–1169 (1995).
Enos, A. P. & Morris, N. R. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell 60, 1019–1027 (1990).
Hagan, I. & Yanagida, M. Novel potential mitotic motor protein encoded by the fission yeast cut7 + gene. Nature 347, 563–566 (1990).
Heck, M. M. et al. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J. Cell Biol. 123, 665–679 (1993).
Kapitein, L. C. et al. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435, 114–118 (2005).
Kashina, A. S. et al. A bipolar kinesin. Nature 379, 270–272 (1996).
Saunders, W. S. & Hoyt, M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70, 451–458 (1992).
Pidoux, A. L., LeDizet, M. & Cande, W. Z. Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol. Biol. Cell 7, 1639–1655 (1996).
Saunders, W., Lengyel, V. & Hoyt, M. A. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell 8, 1025–1033 (1997).
Yukawa, M. & et al. Two spatially distinct kinesin-14 proteins, Pkl1 and Klp2, generate collaborative inward forces against kinesin-5 Cut7 in S. pombe. J. Cell Sci. 131, 210740 (2018).
Mountain, V. et al. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147, 351–366 (1999).
Tanenbaum, M. E., Macurek, L., Galjart, N. & Medema, R. H. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 27, 3235–3245 (2008).
Ferenz, N. P., Paul, R., Fagerstrom, C., Mogilner, A. & Wadsworth, P. Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr. Biol. 19, 1833–1838 (2009).
Florian, S. & Mayer, T. U. The functional antagonism between Eg5 and dynein in spindle bipolarization is not compatible with a simple push-pull model. Cell Rep. 1, 408–416 (2012).
van Heesbeen, R. G., Tanenbaum, M. E. & Medema, R. H. Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep. 8, 948–956 (2014).
Meluh, P. B. & Rose, M. D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell 60, 1029–1041 (1990).
Yukawa, M., Ikebe, C. & Toda, T. The Msd1–Wdr8–Pkl1 complex anchors microtubule minus ends to fission yeast spindle pole bodies. J. Cell Biol. 209, 549–562 (2015).
Decarreau, J. et al. The tetrameric kinesin Kif25 suppresses pre-mitotic centrosome separation to establish proper spindle orientation. Nat. Cell Biol. 19, 384–390 (2017).
Liu, Y. et al. Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol. 37, 314–322 (2019).
Santaguida, S., Vasile, E., White, E. & Amon, A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 29, 2010–2021 (2015).
Mazo, G., Soplop, N., Wang, W. J., Uryu, K. & Tsou, M. F. Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev. Cell 39, 424–437 (2016).
Noda, Y. et al. KIFC3, a microtubule minus end-directed motor for the apical transport of annexin XIIIb-associated Triton-insoluble membranes. J. Cell Biol. 155, 77–88 (2001).
Choi, Y. K., Liu, P., Sze, S. K., Dai, C. & Qi, R. Z. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J. Cell. Biol. 191, 1089–1095 (2010).
Wu, J. et al. Molecular pathway of microtubule organization at the Golgi apparatus. Dev. Cell 39, 44–60 (2016).
Barr, A. R., Kilmartin, J. V. & Gergely, F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J. Cell Biol. 189, 23–39 (2010).
Wang, Z. et al. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J. Biol Chem. 285, 22658–22665 (2010).
Izquierdo, D., Wang, W. J., Uryu, K. & Tsou, M. F. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep. 8, 957–965 (2014).
Acar, S. et al. The bipolar assembly domain of the mitotic motor kinesin-5. Nat. Commun. 4, 1343 (2013).
Magidson, V. et al. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146, 555–567 (2011).
DeBonis, S. et al. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol. Cancer Ther. 3, 1079–1090 (2004).
Skoufias, D. A. et al. S-trityl-l-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 281, 17559–17569 (2006).
Mayer, T. U. et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974 (1999).
Rattner, J. B. & Berns, M. W. Centriole behavior in early mitosis of rat kangaroo cells (PTK2). Chromosoma 54, 387–395 (1976).
Toso, A. et al. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184, 365–372 (2009).
Jean, C., Tollon, Y., Raynaud-Messina, B. & Wright, M. The mammalian interphase centrosome: two independent units maintained together by the dynamics of the microtubule cytoskeleton. Eur. J. Cell Biol. 78, 549–560 (1999).
Meraldi, P. & Nigg, E. A. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J. Cell Sci. 114, 3749–3757 (2001).
Matsuo, K., Nishimura, T., Hayakawa, A., Ono, Y. & Takahashi, M. Involvement of a centrosomal protein kendrin in the maintenance of centrosome cohesion by modulating Nek2A kinase activity. Biochem. Biophys. Res. Commun. 398, 217–223 (2010).
Prosser, S. L., Sahota, N. K., Pelletier, L., Morrison, C. G. & Fry, A. M. Nek5 promotes centrosome integrity in interphase and loss of centrosome cohesion in mitosis. J. Cell Biol. 209, 339–348 (2015).
Kodani, A., Salome Sirerol-Piquer, M., Seol, A., Garcia-Verdugo, J. M. & Reiter, J. F. Kif3a interacts with dynactin subunit p150 glued to organize centriole subdistal appendages. EMBO J. 32, 597–607 (2013).
Konishi, Y. & Setou, M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 12, 559–567 (2009).
Barisic, M. et al. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803 (2015).
Rapley, J. et al. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J. Cell Sci. 121, 3912–3921 (2008).
Eibes, S. et al. Nek9 phosphorylation defines a new role for TPX2 in Eg5-dependent centrosome separation before nuclear envelope breakdown. Curr. Biol. 28, 121–129 (2018). e124.
O’Connell, M. J., Meluh, P. B., Rose, M. D. & Morris, N. R. Suppression of the bimC4 mitotic spindle defect by deletion of klpA, a gene encoding a KAR3-related kinesin-like protein in Aspergillus nidulans. J. Cell Biol. 120, 153–162 (1993).
Sharp, D. J. et al. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell 11, 241–253 (2000).
Prosser, S. L. & Pelletier, L. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 18, 187–201 (2017).
Kuhns, S. et al. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J. Cell Biol. 200, 505–522 (2013).
Kurtulmus, B. et al. LRRC45 contributes to early steps of axoneme extension. J. Cell Sci. 131, 223594 (2018).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Marsh, B. J., Mastronarde, D. N., Buttle, K. F., Howell, K. E. & McIntosh, J. R. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc. Natl Acad. Sci. USA 98, 2399–2406 (2001).
Acknowledgements
We thank A. Neuner and S. Heinze for advice and technical support, B. Kurtulmus for sharing reagents, M. Knop for sharing equipment and helpful discussions, H. Kaessmann for helpful discussions on gene-expression profiles, A. J. Pour and H. Lorenz from the ZMBH Imaging facility for programming plugins and macros used in preliminary data analysis, U. Engel from the Heidelberg University Nikon Imaging Center for support with SIM imaging, M. Langlotz from the ZMBH FACS facility for cell sorting, M. Bornens for the ninein antibody, T. Surrey for the EG5 antibody and J. Pines for the RPE1 FRT/TREx cell line. Electron microscopy and tomography were performed at the Electron Microscopy Core Facility, Heidelberg University. This work was supported by a grant of the German Research Council (Deutsche Forschungsgemeinschaft) DFG-Schi295/6-1 to E.S. and DFG-PE1883-3 and SFB873 to G.P. S.H. was supported by JSPS Research Fellowships for Young Scientists from JSPS, The Naito Grant for studying overseas from The Naito Foundation and Research grant abroad from The Kanae Foundation for the Promotion of Medical Science.
Author information
Authors and Affiliations
Contributions
S.H. and E.S. conceived the project. S.H. designed the study and experiments and performed most of the experiments. A.P.P. performed NEK2-related experiments. M.P. performed preliminary experiments and generated KO cell lines. P.L. performed negative-stain electron microscopy analysis. E.A. performed SIM imaging. C.F. performed electron microscopy tomography. U.J. performed protein purification. G.P. provided reagents and suggestions. S.H. and E.S. wrote the manuscript. All authors contributed to data analysis, discussions and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 KIFC3 regulates MT-based centrosome cohesion.
a, The average of inter-centrosomal distance change in WT and C-NAP1 KO cells during interphase in the live cell imaging shown in Fig. 1a. n = 15 cells each. b, siRNA screen targeting minus-end directed motors as shown in Fig. 1c. Representative images from two independent experiments are shown. Scale bar: 5 μm (1 µm for inset). c, Inter-centrosomal distances from n = 3 biological replicates are plotted (300 cells each) from Fig. 1c and b. d, KIFC3 knockdowns in C-NAP1 KO cells. M, mother centrosome. D, daughter centrosome. Scale bar: 5 μm (1 µm for inset). e, The relative KIFC3 intensity at mother centrosome from d. n = 3 biological replicates, 50 cells each. f, The percentage of cells with separated centrosomes from d. n = 3 biological replicates, 80 cells each. g, Inducible expression of mNG-KIFC3 (siKIFC3 #6 resistant) in C-NAP1 KO cells for 24 hr upon siKIFC3 #6 transfection. Scale bar: 5 μm (1 µm for inset). h, Quantification of g, as in f. n = 3 biological replicates, 80 cells with mNG signal each. i, Inducible expression of rigor mutants in C-NAP1 KO cells. Scale bar: 5 μm (1 µm for inset). j, Quantification of i, as in f. n = 3 biological replicates, 50 cells with mNG signal each. k, NZ treatment for 1 hr after mNG-KIFC3 induction in C-NAP1 KO cells for 24 hr. The percentage of cells with separated centrosomes, as in f. n = 3 biological replicates, 80 cells with mNG signal each. l, The average of inter-centrosomal distance change in C-NAP1 KO cells upon mNG-KIFC3 induction at the middle of the live cell imaging analysis from Fig. 1g. n = 15 cells each. In a, c, e, f, h, j, k and l, data are represented as mean ± s.d. and P values were calculated by Mann–Whitney U test for a and l, and by Two-tailed unpaired Student’s t-test for e, f, h, j and k. Source data for a, c, e, f, h, j, k and l are provided in Supplementary Table 1.
Supplementary Figure 2 Human hKIF25 lacks functional motor domain.
a, RPKM values for hKIF25 and KIFC3 from the RNA-seq dataset of 14 different HeLa cell lines in Liu Y et al.38 (GSE111485). For two HeLa cell lines, mean values of triplicate analyses were used. b, hKIF25 gene lacks N-terminus tail/stalk and a part of the motor domain compared to other primates. The corresponding sequence of the missing part is encoded in the upstream of hKIF25 gene. The non-transcribed part contains 1 bp insertion causing a premature stop codon in the predicted pseudo transcript. c, Comparison of a macaque MfKIF25 (named maKIF25 in figure) transcript (ENSMMUT00000060252.1) and the predicted hKIF25 pseudo transcript. d, Sequencing result of hKIF25 gene, confirming the 1 bp insertion. e, Transfection of hKIF25 siRNAs used in Decarreau J et al.37 in C-Nap1 KO cells. Scale bar: 5 μm (1 µm for inset). f, The percentage of cells with separated centrosomes from e. n = 3 biological replicates, 100 cells each. g, FPKM values for hKIF25 and KIFC3 from the RNA-seq dataset of RPE1 cells in Santaguida S et al.39 (GSE60570). Data for untreated samples in triplicate were used. h, Inducible expression of the indicated construct for 72 hr upon sihKIF25 #2 transfection. Scale bar: 5 μm (1 µm for inset). i, Quantification of h, as in f. n = 3 biological replicates, 80 cells with mNG signal each. j, The relative KIFC3 intensity at mother centrosome from (e). n = 3 biological replicates, 80 cells each. k, KIFC3 protein level in cells transfected with the indicated siRNAs. GM130, loading control. Representative immunoblots from two independent experiments are shown. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. l, Inducible expression of mNG-MfKIF25 (named maKIF25 in figure) for 24 hr. Scale bar: 5 μm (1 µm for inset). m, Quantification of l, as in f. n = 3 biological replicates, 80 cells with mNG signal each. In a, f, g, i, j and m, data are represented as mean ± s.d. and P values were calculated by Two-tailed unpaired Student’s t-test. Source data for a, f, g, i, j and m are provided in Supplementary Table 1.
Supplementary Figure 3 KIFC3 is an outer component of the SDAs.
a, Toroid diameters of the indicated proteins from Fig. 2a are shown. The box and whisker plot shows the median, 25th, and 75th percentiles and minimum and maximum values (n = 50, 25, 25, 50 from left to right, from 25 cells); a representative of two independent experiments. b, Axial distance of the indicated proteins from γ-tubulin PCM pool from Fig. 2a. The box and whisker plot shows the median, 25th, and 75th percentiles and minimum and maximum values (n = 50, 25, 25 from left to right, from 25 cells); a representative of two independent experiments. c, Scheme showing the hierarchy of the SDA components in the mother centrosome. d, Depletion of the SDA components in C-Nap1 KO cells. M, mother centrosome. D, daughter centrosome. Scale bars: 5 µm (1 µm for inset). e, The relative intensities of the SDA components at the mother centrosome during interphase from d. n = 3 biological replicates, >50 cells each. f, Same images in Fig. 2h are shown with different settings of brightness and contrast for mNG-KIFC3 signal. Scale bars: 5 µm (1 µm for inset). g, Protein expression level of mNG-KIFC3 in the condition of Fig. 2h. Representative immunoblots from two independent experiments are shown. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. h, Inducible expression of mNG-KIFC3 in C-Nap1 KO cells upon Dox treatment for 6 hr after Ninein depletion. Representative images from two independent experiments are shown. Scale bar: 5 µm (1 µm for inset). i, The percentage of cells with mNG-KIFC3 on centrosomal MTs from h. n = 3 biological replicates, 100 cells with mNG signal each. In e and i, data are represented as mean ± s.d. and P values were calculated by Two-tailed unpaired Student’s t-test. Source data for a, b, e and i are provided in Supplementary Table 1.
Supplementary Figure 4 Specificity for CDK5RAP2, Myomegalin, PCNT and AKAP450 siRNAs.
a, Protein expression level of CDK5RAP2 in C-Nap1 KO cells transfected with the indicated siRNAs. Representative immunoblots from two independent experiments are shown. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. b, Myomegalin knockdown in RPE1 WT cells. Scale bar: 5 µm (1 µm for inset). c, The relative intensity of Myomegalin in centrosome from b. n = 3 biological replicates, 50 cells each. d, Protein expression level of PCNT in C-Nap1 KO cells transfected with the indicated siRNAs. Representative immunoblots from two independent experiments are shown. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. e, AKAP450 knockdown in RPE1 WT cells. Scale bar: 5 µm (1 µm for inset). f, The relative intensity of AKAP450 in centrosome from e. n = 3 biological replicates, 50 cells each. In c and f, data are represented as mean ± s.d. and P values were calculated by Two-tailed unpaired Student’s t-test. Source data for c and f are provided in Supplementary Table 1.
Supplementary Figure 5 Negative-stain EM analysis of KIFC3.
a, Representative EM micrograph of negative-stained His-SBP-Flag-KIFC3 from two independent experiments are shown. Boxed particles were considered as KIFC3 tetramer. Scale bar: 50 nm. b, The graph shows total length of KIFC3 tetramer from a. Data is represented as mean ± s.d. (n = 20 particles from a single experiment). c, MT cross-linking in C-Nap1 KO cells, from Fig. 4d. The number of crossing points between MTs from the SDAs and the daughter centrosome in a 250 nm thickness section (n = 9 each section from a different cell). Inter-MT distance <24 nm, between the surface of two MTs, was considered as a crossing point. Source data for b and c are provided in Supplementary Table 1.
Supplementary Figure 6 KIFC3 suppresses premature centrosome separation at mitotic onset.
a, Experimental design to classify G2/early prophase cells into early and late G2/early prophase cells. b, Rootletin localization of WT cells in different cell cycle phases. Scale bar: 5 µm. c, The percentage of WT cells with separated centrosomes from b and d. n = 3 biological replicates, 80 cells each. d, NZ treatment for 30 min in the indicated phases as in b. Scale bar: 5 µm. e, Scheme of knocking out KIFC3 gene. f, Detection of KIFC3 KO alleles by genomic PCR. Representative gels from two independent experiments are shown. g, KIFC3 protein level in the indicated cells. Asterisk, a non-specific band. GM-130, loading control. Representative immunoblots from two independent experiments are shown. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. h, Sequencing result of the KIFC3 KO alleles. i, Centrosome separation in KIFC3 KO cells during late G2/early prophase. Scale bar: 5 µm. j, Quantification of i, as in c. n = 3 biological replicates, 60 cells each. k, KIFC3 depletion in WT cells during late G2/early prophase. Scale bar: 5 µm. l, Quantification of k, as in j. n = 3 biological replicates, 90 cells each. m, Effect of mNG-KIFC3 induction for 24 hr on centrosome separation. Scale bar: 5 µm (1 µm for inset). n, The average of inter-centrosomal distance from m. n = 3 biological replicates, >25 cells with mNG signal each. o, Timing of chromosome segregation in WT (n = 139 cells) and KIFC3 KO cells (n = 133 cells) from Fig. 5a, d (n = three independent experiments). p, Chromosome segregation in KIFC3 KO cells. Scale bar: 5 µm. q, The percentage of cells with chromosome mis-segregation from p. n = 3 biological replicates, 80 cells each. In c, j, l, n, o and q, data are represented as mean ± s.d. and P values were calculated by Two-tailed unpaired Student’s t-test for c, j, l, n and q, and by Mann–Whitney U test for o. Source data for c, j, l, n, o and q are provided in Supplementary Table 1.
Supplementary Figure 7 Human hKIF25 does not regulate centrosome cohesion at the onset of mitosis.
a, Scheme of CRISPR/Cas9-mediated large chromosomal deletion in hKIF25 gene. b, Genomic PCR for the detections of WT and KO alleles of the hKIF25 gene. Representative gels from two independent experiments are shown. c, Sequencing result of the hKIFC25 KO alleles. d, Eg5 inhibition in WT and hKIF25 KO cells during early prophase upon 5 µM STLC treatment for 90 min. Scale bar: 5 µm. e, The average of inter-centrosomal distance in early prophase cells from d. n = 3 biological replicates, >30 cells each. f, Quantification of time-lapse imaging of centrosomes in RPE1 WT and hKIF25 KO cells expressing H2B-mNG and γ-tubulin-mRuby2 same as Fig. 5a. Timing of centrosome separation prior to NEBD in WT (n = 121 cells) and hKIF25 KO cells (n = 114 cells) from three independent experiments. g, Inducible expression of mNG-hKIF25 in WT cells during early prophase upon Dox treatment for 24 hr. Scale bar: 5 µm (1 µm for inset). h, Quantification of g, as in e. n = 3 biological replicates, >25 cells each. i, Inducible expression of mNG-MfKIF25 (named maKIF25 in figure) in WT cells during early prophase upon Dox treatment for 24 hr. Scale bar: 5 µm (1 µm for inset). j, Quantification of i, as in e. n = 3 biological replicates, 25 cells each. In e, f, h and j, data are represented as mean ± s.d. and P values were calculated by Two-tailed unpaired Student’s t-test for e, h and j, and by Mann–Whitney U test for f. Source data for e, f, h and j are provided in Supplementary Table 1.
Supplementary Figure 8 NEK2 mediates KIFC3 removal from mitotic centrosomes for centrosome separation in prophase.
a, Scheme of ZFN-mediated knocking out of NEK2 gene. b, Genomic PCR for the detections of WT allele and KO allele in NEK2 gene. Representative gels from two independent experiments are shown. c, Immunoblotting of NEK2 in the lysates of WT cells and the indicated clones. Representative immunoblots from two independent experiments are shown. Scanned images of unprocessed blots are shown in Supplementary Fig. 9. d, NEK2 localization in WT and NEK2 KO clones. Scale bar: 5 µm (1 µm for inset). e, The relative NEK2 intensity at centrosome. n >80 cells each. f, Centrosome separation in WT and NEK2 KO clones during prophase. Scale bar: 5 µm. g, The percentage of prophase cells with separated centrosomes from f. n = 3 biological replicates, ≥40 cells each. h, Overexpression of mNG-NEK2 constructs in RPE1 WT cells. Arrows indicate centrosomes. Scale bar: 5 µm (1 µm for inset). i, The relative intensity of centrosomal KIFC3 in interphase cells from h. n = 3 biological replicates, 50 cells with mNG signal each. j, Metaphase spindle of NEK2 KO cells upon KIFC3 depletion and Aurora A inhibition. Scale bars: 5 µm (1 µm for inset). k, The percentage of metaphase cells with two centrosomes in one pole from j. n = 3 biological replicates, 80 cells each. l, Chromosome segregation of NEK2 KO cells upon KIFC3 depletion and Aurora A inhibition. Arrow indicates a mis-segregated chromosome. Scale bar: 5 µm. m, The percentage of late anaphase/early telophase cells with chromosome mis-segregation from l. n = 3 biological replicates, 100 cells each. In e, g, i, k and m, data are represented as mean ± s.d. and P values were calculated by Two-tailed unpaired Student’s t-test. Source data for g, i, k and m are provided in Supplementary Table 1.
Supplementary Figure 9
Scans of unprocessed Western blots and CBB stain.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and legends for Supplementary Videos 1–6 and Supplementary Table 1.
Supplementary Table 1
Source data
Supplementary Video 1
Supplementary Video 1
Supplementary Video 2
Supplementary Video 2
Supplementary Video 3
Supplementary Video 3
Supplementary Video 4
Supplementary Video 4
Supplementary Video 5
Supplementary Video 5
Supplementary Video 6
Supplementary Video 6
Rights and permissions
About this article
Cite this article
Hata, S., Pastor Peidro, A., Panic, M. et al. The balance between KIFC3 and EG5 tetrameric kinesins controls the onset of mitotic spindle assembly. Nat Cell Biol 21, 1138–1151 (2019). https://doi.org/10.1038/s41556-019-0382-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0382-6
This article is cited by
-
Kinesin KIFC3 is essential for microtubule stability and cytokinesis in oocyte meiosis
Cell Communication and Signaling (2024)
-
Molecular basis promoting centriole triplet microtubule assembly
Nature Communications (2024)
-
An antioxidant screen identifies ascorbic acid for prevention of light-induced mitotic prolongation in live cell imaging
Communications Biology (2023)
-
KIF11 serves as a cell cycle mediator in childhood acute lymphoblastic leukemia
Journal of Cancer Research and Clinical Oncology (2023)
-
NAT10 regulates mitotic cell fate by acetylating Eg5 to control bipolar spindle assembly and chromosome segregation
Cell Death & Differentiation (2022)