Abstract
Ca2+/calmodulin-dependent kinase II (CaMKII) is a multifunctional serine/threonine kinase family, and its δ isoform is predominant in the heart. Excessive CaMKII activation plays a pivotal role in the pathogenesis of severe heart conditions, including myocardial infarction, cardiomyopathy and heart failure. However, the identity of CaMKII splice variants and the mechanism(s) underlying CaMKII-mediated cardiac pathology remain elusive. Here, we show that CaMKII-δ9, the most abundant CaMKII-δ splice variant in human heart, potently promotes cardiomyocyte death, cardiomyopathy and heart failure by disrupting cardiomyocyte genome stability. Mechanistically, CaMKII-δ9, but not the previously well-studied CaMKII-δ2 and CaMKII-δ3, targets the ubiquitin-conjugating enzyme E2T (UBE2T) for phosphorylation and degradation, disrupting UBE2T-dependent DNA repair and leading to the accumulation of DNA damage and genome instability. These findings not only reveal a crucial role of CaMKII in the regulation of DNA repair, but also mark the CaMKII-δ9–UBE2T–DNA damage pathway as an important therapeutic target for cardiomyopathy and heart failure.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All of the sequencing data that support the findings of this study have been deposited in the NCBI under accession codes PRJNA381326, PRJNA381328 and PRJNA381329. Previously published Deep-seq data that were re-analysed here are available under accession codes SRR830965, SRR830966, SRR830967, SRR830968, SRR830969, SRR830970, SRR830971, SRR830972, SRX196319, SRX196328, SRX196337, SRX081927, SRX081928, SRX494639, SRX066573, SRR1735880, SRR1735881, SRR1735882, SRR1735883, SRR1735884, SRR1735885, SRX471444, SRX471445, SRX471446, SRX471447, SRX471460, SRX471461 and SRX471462. All of the source data have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
Campisi, J. & d’Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 (2007).
Schreader, B. A. & Nambu, J. R. A fine balance for life and death decisions. Nat. Struct. Mol. Biol. 11, 386–388 (2004).
Roos, W. P., Thomas, A. D. & Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 16, 20–33 (2016).
Tubbs, A. & Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017).
Wang, J. & Lindahl, T. Maintenance of genome stability. Gen. Proteom. Bioinf. 14, 119–121 (2016).
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2016).
Shukla, P. C., Singh, K. K., Yanagawa, B., Teoh, H. & Verma, S. DNA damage repair and cardiovascular diseases. Can. J. Cardiol. 26, 13A–16A (2010).
Shukla, P. C. et al. BRCA1 is an essential regulator of heart function and survival following myocardial infarction. Nat. Commun. 2, 593 (2011).
Higo, T. et al. DNA single-strand break-induced DNA damage response causes heart failure. Nat. Commun. 8, 15104 (2017).
Erickson, J. R., He, B. J., Grumbach, I. M. & Anderson, M. E. CaMKII in the cardiovascular system: sensing redox states. Physiol. Rev. 91, 889–915 (2011).
Zhu, W. Z. et al. Linkage of beta1-adrenergic stimulation to apoptotic heart cell death through protein kinase A-independent activation of Ca2+/calmodulin kinase II. J. Clin. Investig. 111, 617–625 (2003).
Peng, W. et al. Cardioprotection by CaMKII-δB is mediated by phosphorylation of heat shock factor 1 and subsequent expression of inducible heat shock protein 70. Circ. Res. 106, 102–110 (2010).
Mayer, P., Mohlig, M., Schatz, H. & Pfeiffer, A. Additional isoforms of multifunctional calcium/calmodulin-dependent protein kinase II in rat heart tissue. Biochem. J. 298, 757–758 (1994).
Mayer, P., Mohlig, M., Idlibe, D. & Pfeiffer, A. Novel and uncommon isoforms of the calcium sensing enzyme calcium/calmodulin dependent protein kinase II in heart tissue. Basic Res. Cardiol. 90, 372–379 (1995).
Hoch, B., Meyer, R., Hetzer, R., Krause, E. G. & Karczewski, P. Identification and expression of δ-isoforms of the multifunctional Ca2+/calmodulin-dependent protein kinase in failing and nonfailing human myocardium. Circ. Res. 84, 713–721 (1999).
Edman, C. F. & Schulman, H. Identification and characterization of δ B-CaM kinase and δ C-CaM kinase from rat heart, two new multifunctional Ca2+/calmodulin-dependent protein kinase isoforms. Biochim. Biophys. Acta 1221, 89–101 (1994).
Mishra, S., Gray, C. B., Miyamoto, S., Bers, D. M. & Brown, J. H. Location matters: clarifying the concept of nuclear and cytosolic CaMKII subtypes. Circ. Res. 109, 1354–1362 (2011).
Hagemann, D., Bohlender, J., Hoch, B., Krause, E. G. & Karczewski, P. Expression of Ca2+/calmodulin-dependent protein kinase II δ-subunit isoforms in rats with hypertensive cardiac hypertrophy. Mol. Cell. Biochem. 220, 69–76 (2001).
Little, G. H. et al. Critical role of nuclear calcium/calmodulin-dependent protein kinase IIδB in cardiomyocyte survival in cardiomyopathy. J. Biol. Chem. 284, 24857–24868 (2009).
Gray, C. B. et al. CaMKIIδ subtypes differentially regulate infarct formation following ex vivo myocardial ischemia/reperfusion through NF-kappaB and TNF- α. J. Mol. Cell. Cardiol. 103, 48–55 (2017).
Zhang, T. et al. CaMKII is a RIP3 substrate mediating ischemia- and oxidative stress-induced myocardial necroptosis. Nat. Med. 22, 175–182 (2016).
Joiner, M. L. et al. CaMKII determines mitochondrial stress responses in heart. Nature 491, 269–273 (2012).
He, B. J. et al. Oxidation of CaMKII determines the cardiotoxic effects of aldosterone. Nat. Med. 17, 1610–1618 (2011).
Zhang, R. et al. Calmodulin kinase II inhibition protects against structural heart disease. Nat. Med. 11, 409–417 (2005).
van Oort, R. J. et al. Ryanodine receptor phosphorylation by calcium/calmodulin-dependent protein kinase II promotes life-threatening ventricular arrhythmias in mice with heart failure. Circulation 122, 2669–2679 (2010).
Mesubi, O. O. & Anderson, M. E. Atrial remodelling in atrial fibrillation: CaMKII as a nodal proarrhythmic signal. Cardiovasc. Res. 109, 542–557 (2016).
Zhang, T. et al. The δC isoform of CaMKII is activated in cardiac hypertrophy and induces dilated cardiomyopathy and heart failure. Circ. Res. 92, 912–919 (2003).
Ling, H. et al. Requirement for Ca2+/calmodulin-dependent kinase II in the transition from pressure overload-induced cardiac hypertrophy to heart failure in mice. J. Clin. Investig. 119, 1230–1240 (2009).
Sharon, D., Tilgner, H., Grubert, F. & Snyder, M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 31, 1009–1014 (2013).
Gerber, S. A., Rush, J., Stemman, O., Kirschner, M. W. & Gygi, S. P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl Acad. Sci. USA 100, 6940–6945 (2003).
Kawakami, H. et al. Simultaneous absolute quantification of 11 cytochrome P450 isoforms in human liver microsomes by liquid chromatography tandem mass spectrometry with in silico target peptide selection. J. Pharm. Sci. 100, 341–352 (2011).
Erickson, J. R. et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 133, 462–474 (2008).
Machida, Y. J. et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23, 589–596 (2006).
Rickman, K. A. et al. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep. 12, 35–41 (2015).
Bartek, J. & Lukas, J. Balancing life-or-death decisions. Science 314, 261–262 (2006).
Roos, W. P. & Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12, 440–450 (2006).
Kuo, L. J. & Yang, L. X. γ-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo 22, 305–309 (2008).
Olive, P. L. & Banath, J. P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 1, 23–29 (2006).
Niedernhofer, L. J., Lalai, A. S. & Hoeijmakers, J. H. Fanconi anemia (cross)linked to DNA repair. Cell 123, 1191–1198 (2005).
Ceccaldi, R., Sarangi, P. & D’Andrea, A. D. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 17, 337–349 (2016).
Donahue, S. L. & Campbell, C. A DNA double strand break repair defect in Fanconi anemia fibroblasts. J. Biol. Chem. 277, 46243–46247 (2002).
Richardson, C. D. et al. CRISPR–Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat. Genet. 50, 1132–1139 (2018).
Wang, X. et al. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 27, 3098–3108 (2007).
Zhu, W. et al. Activation of CaMKIIδC is a common intermediate of diverse death stimuli-induced heart muscle cell apoptosis. J. Biol. Chem. 282, 10833–10839 (2007).
Ling, H. et al. Ca2+/Calmodulin-dependent protein kinase II δ mediates myocardial ischemia/reperfusion injury through nuclear factor-kappaB. Circ. Res. 112, 935–944 (2013).
Singh, M. V. et al. Ca2+/calmodulin-dependent kinase II triggers cell membrane injury by inducing complement factor B gene expression in the mouse heart. J Clinical Investig. 119, 986–996 (2009).
Schwab, R. A. et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol. Cell 60, 351–361 (2015).
Zhang, X. et al. Rhesus macaques develop metabolic syndrome with reversible vascular dysfunction responsive to pioglitazone. Circulation 124, 77–86 (2011).
Treutlein, B., Gokce, O., Quake, S. R. & Sudhof, T. C. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc. Natl Acad. Sci. USA 111, E1291–E1299 (2014).
Liu, F. et al. Upregulation of MG53 induces diabetic cardiomyopathy through transcriptional activation of peroxisome proliferation-activated receptor α. Circulation 131, 795–804 (2015).
Burridge, P. W. et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 11, 855–860 (2014).
Acknowledgements
We thank H. Cheng and I. C. Bruce for constructive comments on the manuscript, and H. Shang, W. Zheng, Y. Liu, D. Ma, Y. Wang, P. Xie and X. Wang for their excellent technical support. This work was supported by the National Key R&D Program of China (2018YFA0800501 and 2018YFA0507603), the National Natural Science Foundation of China (31671177, 81630008, 81790621, 31521062 and 81370234), the Beijing Municipal Science & Technology Commission (Z171100000417006) and the Beijing Natural Science Foundation (5182010). The normal human tissue was obtained from the NIH NeuroBioBank at the University of Maryland, Baltimore, MD.
Author information
Authors and Affiliations
Contributions
M.Z. and Y.Z. generated the initial idea and conducted key experiments. M.Z., Y.Z. and R.-P.X. proposed the hypothesis, designed the study, supervised the experiments and data analysis, wrote the manuscript and interpreted the results. H.G., D.L., X.S., L.J., Y.L., Y.T., Y.S., J.L., X.H., L.S., J.Q., F.W., F.L. and R.-P.X. performed experiments. X.Z., P.Y. and C.-Y.L. helped with the bioinformatics analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 CaMKII-δ9 is present in the heart.
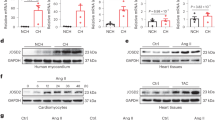
a, Schematic of CaMKII-δ splice variants. CaMKII-δ mainly undergoes alternative splicing events at two variable domains, one between exons 13 and 17, and the other after exon 20. Exons are numbered, and full-size boxes represent coding exons, while smaller, green boxes stand for untranslated regions (UTRs), with special exons colored. b, Strategy for SMRT sequencing of full-length CaMKII-δ transcripts. CaMKII-δ transcripts were reverse-transcribed from total RNA isolated from the hearts of different species. Each cDNA was amplified with paired primers, whose locations are marked with arrows of different colors. The forward primer (red) is located on exon 1, and the reverse primer (blue) is located on exon 22. The PCR products were concentrated and purified for SMRT sequencing. c-e, Percentages of exon junctions of CaMKII-δ assayed by RNA-seq in variable domain 1 (between exons 13 and 17 (c) and 14-17 (d)), and variable domain 2 (between exons 20 and 22) (e) in the hearts of human, rhesus monkey, dog, rat, and mouse. In panel e, in the hearts of monkey and human, exon 20b is another form of exon 20, which is 147 bases longer than the classic exon 20, and has not been described before. Data are mean ± s.e.m.. n = 8 (human and rat), 7 (rhesus monkey), and 6 (dog and mouse) biologically independent samples. Data are mean ± s.e.m.. One-way ANOVA. The source data for the graphs are in Supplementary Table 5.
Supplementary Figure 2 Identification of CaMKII-δ9 protein in the heart.
a, Peptides sequences of exons 16 and 21 used as antigens for the production of antibodies. b, Immunoblots of NRVMs transfected with Ad-β-gal, Ad-HA-CaMKII-δ2, Ad-HA-CaMKII-δ3, or Ad-Flag-CaMKII-δ9, with serum containing anti-exon 16 or anti-exon 21. Data shown represent 3 independent experiments. c, d, Western blots of Flag-tagged CaMKII-δ9 recombinant protein with anti-exon 16 (c) and anti-exon 21 (d) with increasing ratios of the corresponding antigen peptide to CaMKII-δ9 protein. n = 4 biologically independent samples. Mean ± s.e.m.. One-way ANOVA. e, f, Heart lysates of 10-week old mice immunoprecipitated with anti-exon 21 (e) or anti-exon 16 (f), followed by SDS-PAGE and Coomassie blue staining. The bands at ~50 kD (red boxes) were cut for MS analysis. g, h, LC-MS/MS spectra of the peptides matching the junction of CaMKII-δ exons 13-16-17 (g) and exons 20-21 (h) from mouse hearts immunoprecipitated with anti-exon 21 (g) or anti-exon 16 (h). The peptide sequence above is the corresponding exon junction, with the specific exons 16 and 21 in bold. bn ions (red) are fragments at the nth peptide bonds that contain the amino-terminal part of the peptide, whereas yn ions (blue) contain the carboxy-terminal part. NL, normalized intensity level (counts per second). i, j, CaMKII-δ9 tissue distribution in rhesus monkeys (i) and wt mice (j). n = 4 biologically independent samples. CaMKII-δ9 recombinant protein served as a positive control (PC). In both species, a band with a higher molecular weight was detected in the brain, which was the brain-enriched splice variant CaMKII-δ1 (exons 13-15-16-17). k, l, Immunofluorescent confocal microscopic images of the cytosolic location of endogenous CaMKII-δ9 (green) in adult (left) and neonatal (right) rat ventricular cardiomyocytes (k, n = 4 biologically independent samples), and the nuclear location of HA-CaMKII-δ3 (red) in NRVMs infected with Ad-HA-CaMKII-δ3 (l, Data shown represent 6 independent experiments). Scale bars, 10 μm. m, CaMKII-δ9 protein levels in nuclear and cytosolic fractions of NRVMs. Data shown represent 6 independent experiments. The source data for the graphs are in Supplementary Table 5 and unprocessed blots in Supplementary Fig. 9.
Supplementary Figure 3 Pathological relevance of CaMKII-δ9 in the heart.
a, b, Western blots showing the phosphorylation (a, n = 8 biologically independent samples) and oxidation (b, n = 7 biologically independent samples) levels of CaMKII-δ9 in NRVMs with or without Dox treatment (1 μM, 30 and 60 min). The NRVMs were infected with Ad-Flag-CaMKII-δ9, and the lysates were immunoprecipitated with Flag antibody and subjected to western blot analysis. c, d, Representative western blots and statistical data showing the phosphorylation (c) and oxidation (d) levels of CaMKII-δ9 in perfused mouse hearts with or without I/R injury (30 min ischemia followed by 60 min reperfusion). n = 6 biologically independent samples. The heart lysates were immunoprecipitated with exon 16 antibody and subjected to western blot analysis. e, Sequence of CaMKII-δ9 siRNA. The black sequence is the siRNA target in exon 16 of CaMKII-δ9. The siRNA sequence is below in red. f, g, Knockdown efficiency of CaMKII-δ9 siRNA confirmed by mRNA (f, n = 5 (scrambled), and 8 (CaMKII-δ9 siRNA) biologically independent samples) and protein (g, n = 3 biologically independent samples) levels. h, Averaged data of mRNA levels of CaMKII-δ2 and CaMKII-δ3 assayed by real-time PCR in NRVMs infected with scrambled or CaMKII-δ9 siRNA. n = 18 (CaMKII-δ2), and 15 (CaMKII-δ3) biologically independent samples. i, j, Cell viability assessed by LDH concentration in the culture medium of NRVMs treated with scrambled or CaMKII-δ9 siRNAs with or without H2O2 (200 μM) (i) or Dox (1 μM) (j). n = 6 biologically independent samples. k, Viability assessed by LDH in the medium of NRVMs infected with Ad-β-gal, Ad-CaMKII-δ9, and Ad-CaMKII-δ2 at the indicated MOI for 48 h. n = 10 biologically independent samples. l, Representative western blots and statistical data of CaMKII-δ expression in NRVMs infected with Ad-β-gal, Ad-CaMKII-δ9, or Ad-CaMKII-δ2 (MOI 50, 48 h). n = 3 biologically independent samples. Data are mean ± s.e.m.. One-way ANOVA (a, b, g, k), two-sided Student’s t-test (c, d, f, h, l), or two-way ANOVA (i, j). The source data for the graphs are in Supplementary Table 5 and unprocessed blots in Supplementary Fig. 9.
Supplementary Figure 4 RNA-seq analysis of the gene expression profiles of CaMKII-δ9 and CaMKII-δ2.
a, Heatmap representing the gene expression signature among NRVMs infected with Ad-β-gal, Ad-CaMKII-δ9, or Ad-CaMKII-δ2 (MOI 50, 48 h) based on genes differentially-expressed between Ad-β-gal and Ad-CaMKII-δ9. The expression value of each gene was calculated as fragments per kilobase of transcript, per million mapped fragments (FPKM). Seventy-seven genes are listed; they were differentially and significantly changed (>1.5-fold or <0.67-fold, n = 3 biologically independent samples) in cells infected with Ad-CaMKII-δ9 relative to those in cells infected with Ad-β-gal. The 15 genes that were different regulated in Ad-CaMKII-δ9 and Ad-CaMKII-δ2 are in red. The heatmap was generated by the R package “pheatmap” with the option “scale = row”, which means the expression value of each gene is the z-score normalized by the FPKM value. b, Data of mRNA levels assayed by real-time PCR of 12 of the 15 genes identified by RNA-seq to be regulated by CaMKII-δ9, but not CaMKII-δ2, in NRVMs infected with Ad-β-gal, Ad-CaMKII-δ9, or Ad-CaMKII-δ2 (MOI 50, 48 h). n = 14 biologically independent samples. c, d, Protein levels of COX-2 (c) and PAI-2 (d) in cultured NRVMs infected with Ad-β-gal, Ad-CaMKII-δ9, or Ad-CaMKII-δ2 (dose as indicated, 48 h). n = 4 biologically independent samples. e, Typical western blots and averaged data showing the protein level of COX-2 in NRVMs transfected with scrambled or COX-2 siRNAs. n = 4 biologically independent samples. f, g, Cell viability indexed by caspase 3/7 activity (f) and LDH concentration in the culture medium (g) in NRVMs infected with Ad-β-gal or Ad-CaMKII-δ9 in the presence or absence of COX-2 siRNAs. n = 3 (f), and 6(g) biologically independent samples. h, Representative western blots and averaged data showing the protein levels of UBE2T in NRVMs transfected with scrambled or UBE2T siRNAs. n = 4 biologically independent samples. Data are mean ± s.e.m.. One-way ANOVA (b, c, d, e, h), or two-way ANOVA (f, g). The source data for the graphs are in Supplementary Table 5 and unprocessed blots in Supplementary Fig. 9.
Supplementary Figure 5 CaMKII-δ9 induced cardiomyocyte DNA damage.
a, b, Complete representative immunostaining of γH2AX (a) and comet assays (b) in NRVMs infected with Ad-β-gal, Ad-CaMKII-δ2, or Ad-CaMKII-δ9 (MOI 50, 48 h). Scale bars, 20 μm. Part of the representative images and averaged data are shown in Fig. 3a, c. c-f, Representative western blots and statistical data showing the levels of FANCD2 (c, n = 4 biologically independent samples) and FANCI (d, n = 4 biologically independent samples), and caspase 3/7 activity (e, f, n = 5 biologically independent samples) in NRVMs infected with scrambled, FANCD2 or FANCI siRNAs. Data are mean ± s.e.m.. One-way ANOVA. The source data for the graphs are in Supplementary Table 5 and unprocessed blots in Supplementary Fig. 9.
Supplementary Figure 6 Role of CaMKII-δ9-UBE2T signaling in cardiac pathophysiology.
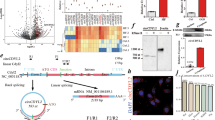
a, Schematic of CaMKII-δ9 tg mouse construction. b, PCR genotyping of wt and CaMKII-δ9 tg mice. The genotyping was independently repeated 6 times. c, d, Ratio of heart weight to body weight (c, n =15 (wt), and 9 (CaMKII-δ9 tg) biologically independent animals) and ventricular gene expression (d, n = 7 (wt), and 6 (CaMKII-δ9 tg) biologically independent animals) of wt and CaMKII-δ9 tg mice at 10-week age. e, Immunostaining of cardiac γH2AX of wt and CaMKII-δ9 tg mice (10 weeks of age). Arrows, γH2AX-positive cells. Right window, enlarged image of the view as indicated. Averaged data are in Fig. 4g. Scale bar, 20 μM. f, Schematic of CaMKII-δ9 shRNA transgenic (shRNA tg) mouse construction. g, PCR genotyping of wt and shRNA tg mice. The genotyping was independently repeated 6 times. h, Cardiac exon 21 protein levels of wt and shRNA tg mice (10 weeks age) (n =4 biologically independent animals). i, j, Ratio of heart weight to body weight (i, n = 16 (wt), and 8 (shRNA tg) biologically independent animals), and cardiac UBE2T protein levels (j, n = 4 biologically independent animals) of wt and shRNA tg mice 4 weeks after TAC surgery. k, Cardiac CaMKII-δ protein levels of CaMKII-δ9 tg and CaMKII-δ2 tg mice at the age of 10 weeks (n = 4 biologically independent animals). l-q, Cardiac TUNEL staining (l, n = 8 biologically independent animals), echocardiography (m, n = 8 biologically independent animals), ratio of heart weight to body weight (n, n = 8 biologically independent animals), Kaplan-Meier survival curves (o, n = 10 (wt), 19 (CaMKII-δ9 tg), and 12 (CaMKII-δ2 tg) biologically independent animals), cardiac γH2AX staining (p, n = 8 biologically independent animals), and cardiac UBE2T protein levels (q, n = 4 biologically independent animals) from wt, CaMKII-δ9 tg and CaMKII-δ2 tg mice. Data are mean ± s.e.m.. Two-sided Student’s t-test (c, d, i), one-way ANOVA (k-n, p, q), two-way ANOVA (j), or log-rank (Mantel-Cox) test (o). The source data for the graphs are in Supplementary Table 5 and unprocessed blots in Supplementary Fig. 9.
Supplementary Figure 7 CaMKII-δ9 phosphorylates UBE2T at Ser110 site.
a, Mass spectrometric data showing two potential phosphorylation sites (Ser110 and Ser193, red circles) of UBE2T mediated by CaMKII-δ9 (n = 3 biologically independent samples). HEK293 cells were transfected with myc-tagged UBE2T in the presence of control vector or Flag-tagged CaMKII-δ9, whole-cell lysates were immunoprecipitated with myc antibody, and then the immuno-complex was subjected to post-translational modification mass spectrometric analysis. b, Sequence alignment of UBE2T protein showing conservation of the Ser110 site, but not the Ser193 site of UBE2T (red arrows) in 12 species. c, Typical western blots and averaged data showing the serine phosphorylation and total levels of UBE2T and UBE2T-S110A recombinant protein with or without CaMKII-δ9 protein co-incubation in a cell-free system (n = 4 biologically independent samples). Two-way ANOVA. d, Representative immunofluorescence images of wt UBE2T, UBE2T-S110A, and UBE2T-S193A (all with myc tag) in NRVMs infected with Ad-UBE2T, Ad-UBE2T-S110A, or Ad-UBE2T-S193A (Data shown represent 6 independent experiments). Note that UBE2T-S110A was resistant to degradation, and distributed in both cytoplasm and nucleus, indicating that the Ser110 site of UBE2T is responsible for its degradation, but not its subcellular distribution, Scale bar, 10 μm. Data are mean ± s.e.m.. The source data for the graphs are in Supplementary Table 5 and unprocessed blots in Supplementary Fig. 9.
Supplementary Figure 8 Interaction between the peptides encoded by CaMKII-δ exon junctions and UBE2T.
a-c, Co-immunoprecipitation of the peptides encoded by CaMKII-δ exon junctions of exons 13-17 (a), 13-14-17 (b), and 13-15-16-17 (c), with UBE2T in HEK293 cells transfected with plasmids of the corresponding exon junctions (tagged with Flag-GFP), or UBE2T-myc. (Data shown represent 4 independent experiments). The unprocessed blots in Supplementary Fig. 9.
Supplementary Figure 9 Uncropped images from Western blots.
Uncropped images for Fig. 1b, e-h and Fig. 2e, f, i. Uncropped images for Fig. 3b, l, m, Fig. 4a, h, i, Fig. 5f, and Fig. 6a–c, e, f, h, i. Uncropped images for Fig. 7a, b, d–i. Uncropped images for Fig. 8a–e. Uncropped images for Supplementary Fig. 2b-d,i,j,m. Uncropped images for Supplementary Fig. 3a-d,g,l, and Supplementary Fig. 4c-e,h. Uncropped images for Supplementary Fig. 5c,d, Supplementary Fig. 6h,j,k,q and Supplementary Fig. 7c. Uncropped images for Supplementary Fig. 8a-c.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and their legends and legends for Supplementary Tables 1–5.
Supplementary Table 1
Identified proteotypic trypsin peptides for quantitative MS in the hearts.
Supplementary Table 2
Gene expression profiles of cardiomyocytes infected with Ad-CaMKII-δ9 or Ad-CaMKII-δ2 over Ad-β-gal.
Supplementary Table 3
The primer pairs used for quantitative real-time PCR analysis of gene expression.
Supplementary Table 4
The target sequences of siRNAs.
Supplementary Table 5
Statistics source data.
Rights and permissions
About this article
Cite this article
Zhang, M., Gao, H., Liu, D. et al. CaMKII-δ9 promotes cardiomyopathy through disrupting UBE2T-dependent DNA repair. Nat Cell Biol 21, 1152–1163 (2019). https://doi.org/10.1038/s41556-019-0380-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0380-8
This article is cited by
-
Cigarette tar accelerates atherosclerosis progression via RIPK3-dependent necroptosis mediated by endoplasmic reticulum stress in vascular smooth muscle cells
Cell Communication and Signaling (2024)
-
SNTA1-deficient human cardiomyocytes demonstrate hypertrophic phenotype and calcium handling disorder
Stem Cell Research & Therapy (2022)
-
CDC-like kinase 4 deficiency contributes to pathological cardiac hypertrophy by modulating NEXN phosphorylation
Nature Communications (2022)
-
RBM24 controls cardiac QT interval through CaMKIIδ splicing
Cellular and Molecular Life Sciences (2022)
-
Mitochondrial CaMKII causes adverse metabolic reprogramming and dilated cardiomyopathy
Nature Communications (2020)