Abstract
Migrasomes are recently discovered cellular organelles that form as large vesicle-like structures on retraction fibres of migrating cells. While the process of migrasome formation has been described before, the molecular mechanism underlying migrasome biogenesis remains unclear. Here, we propose that the mechanism of migrasome formation consists of the assembly of tetraspanin- and cholesterol-enriched membrane microdomains into micron-scale macrodomains, which swell into migrasomes. The major finding underlying the mechanism is that tetraspanins and cholesterol are necessary and sufficient for migrasome formation. We demonstrate the necessity of tetraspanins and cholesterol via live-cell experiments, and their sufficiency by generating migrasome-like structures in reconstituted membrane systems. We substantiate the mechanism by a theoretical model proposing that the key factor driving migrasome formation is the elevated membrane stiffness of the tetraspanin- and cholesterol-enriched macrodomains. Finally, the theoretical model was quantitatively validated by experimental demonstration of the membrane-stiffening effect of tetraspanin 4 and cholesterol.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Code availability
The main computational codes used for the simulations in this paper are available at https://github.com/benzucker-tau/Migrasome-simulation. Further codes in surface evolver and codes in Matlab used for analyses are available from the corresponding authors upon reasonable request.
Change history
21 August 2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Taylor, A. C. & Robbins, E. Observations on microextensions from the surface of isolated vertebrate cells. Dev. Biol. 6, 660–673 (1963).
Ma, L. et al. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 25, 24–38 (2015).
Wu, D. et al. Pairing of integrins with ECM proteins determines migrasome formation. Cell Res. 27, 1397–1400 (2017).
Jiang, D. et al. Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat. Cell Biol. https://doi.org/10.1038/s41556-019-0358-6 (2019).
Rubinstein, E. The complexity of tetraspanins. Biochem. Soc. Trans. 39, 501–505 (2011).
Charrin, S. et al. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154 (2009).
Zuidscherwoude, M. et al. The tetraspanin web revisited by super-resolution microscopy. Sci. Rep. 5, 12201 (2015).
Le Naour, F., Andre, M., Boucheix, C. & Rubinstein, E. Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics 6, 6447–6454 (2006).
Hemler, M. E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Ann. Rev. Cell Dev. Biol. 19, 397–422 (2003).
Borghi, N., Rossier, O. & Brochard-Wyart, F. Hydrodynamic extrusion of tubes from giant vesicles. Europhys. Lett. 64, 837–843 (2003).
Rossier, O. et al. Giant vesicles under flows: extrusion and retraction of tubes. Langmuir 19, 575–584 (2003).
Solon, J., Streicher, P., Richter, R., Brochard-Wyart, F. & Bassereau, P. Vesicles surfing on a lipid bilayer: self-induced haptotactic motion. Proc. Natl Acad. Sci. USA 103, 12382–12387 (2006).
Delorme, N. & Fery, A. Direct method to study membrane rigidity of small vesicles based on atomic force microscope force spectroscopy. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 74, 030901 (2006).
Landau, L. D. & Lifshitz, E. M. Theory of Elasticity 1st edn, Vol. 7 (Butterworth-Heinemann, 1959).
Dimova, R. Recent developments in the field of bending rigidity measurements on membranes. Adv. Colloid Interface Sci. 208, 225–234 (2014).
Usery, R. D. et al. Membrane bending moduli of coexisting liquid phases containing transmembrane peptide. Biophys. J. 115, 164 (2018).
Oradd, G., Shahedi, V. & Lindblom, G. Effect of sterol structure on the bending rigidity of lipid membranes: a 2H NMR transverse relaxation study. Biochim. Biophys. Acta 1788, 1762–1771 (2009).
Aimon, S. et al. Membrane shape modulates transmembrane protein distribution. Dev. Cell 28, 212–218 (2014).
Tian, A., Johnson, C., Wang, W. & Baumgart, T. Line tension at fluid membrane domain boundaries measured by micropipette aspiration. Phys. Rev. Lett. 98, 208102 (2007).
Kuzmin, P. I., Akimov, S. A., Chizmadzhev, Y. A., Zimmerberg, J. & Cohen, F. S. Line tension and interaction energies of membrane rafts calculated from lipid splay and tilt. Biophys. J. 88, 1120–1133 (2005).
Baumgart, T., Das, S., Webb, W. W. & Jenkins, J. T. Membrane elasticity in giant vesicles with fluid phase coexistence. Biophys. J. 89, 1067–1080 (2005).
Bar-Ziv, R. & Moses, E. Instability and “pearling” states produced in tubular membranes by competition of curvature and tension. Phys. Rev. Lett. 73, 1392–1395 (1994).
Tsafrir, I. et al. Pearling instabilities of membrane tubes with anchored polymers. Phys. Rev. Lett. 86, 1138–1141 (2001).
Bar-Ziv, R., Tlusty, T., Moses, E., Safran, S. A. & Bershadsky, A. Pearling in cells: a clue to understanding cell shape. Proc. Natl Acad. Sci. USA 96, 10140–10145 (1999).
Su, Q. P. et al. Vesicle size regulates nanotube formation in the cell. Sci. Rep. 6, 24002 (2016).
Du, W. et al. Kinesin 1 drives autolysosome tubulation. Dev. Cell 37, 326–336 (2016).
Wang, C. et al. Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res. 25, 1108–1120 (2015).
Martin, B. J. & van Golen, K. L. A comparison of cholesterol uptake and storage in inflammatory and noninflammatory breast cancer cells. Int. J. Breast Cancer 2012, 412581 (2012).
Renaud, J. F. et al. Normal serum and lipoprotein-deficient serum give different expressions of excitability, corresponding to different stages of differentiation, in chicken cardiac cells in culture. Proc. Natl Acad. Sci. USA 79, 7768–7772 (1982).
Wang, X. & Seed, B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31, e154 (2003).
Spandidos, A. et al. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics 9, 633 (2008).
Spandidos, A., Wang, X., Wang, H. & Seed, B. Primer bank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 (2010).
Wang, X., Spandidos, A., Wang, H. & Seed, B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40, D1144–D1149 (2012).
Acknowledgements
The authors are grateful to members of the Kozlov and Yu groups for helpful discussions. This research was supported by the Ministry of Science and Technology of the People’s Republic of China (2016YFA0500202 and 2017YFA0503404) to L.Y., the National Natural Science Foundation of China (31430053 and 31621063), the Natural Science Foundation of China international cooperation and exchange program (31561143002), the Independent Research of Tsinghua University (20161080135) to L.Y., and the Israel Science Foundation grant 1066/15, EU consortium InCeM to M.M.K., who also holds the Joseph Klafter Chair in Biophysics.
Author information
Authors and Affiliations
Contributions
L.Y., Y.H., S.Z., Y.S. and J.L. conceived the experiments. M.M.K. and B.Z. conceived the modelling. L.Y. and M.M.K. wrote the paper and supervised the project. Y.H., S.Z., Y.Z., H.C., T.D. and Y.L. carried out the experiments. B.Z. and S.E. performed the computations. All authors discussed the manuscript, commented on the project and contributed to preparing the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
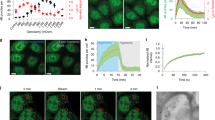
Supplementary Figure 1 Tspan4 is necessary for migrasome formation.
a, mRNA levels of Tspan4 in Tspan4-GFP-expressing NRK cell lines and their parent cells were measured by real-time PCR. Data shown represent mean±SEM (n=3 independent experiments). b, Cells with stable Tspan4-GFP expression were treated with or without deglycosylation mix, then lysed and analyzed by western blot. c, mRNA levels of 9 TSPAN genes in the MGC-803 cell line were measured by real-time PCR and the values were normalized by the TSPAN4 mRNA level. Data shown represent mean±SEM (n=3 independent experiments). d, Verification of the NRK Tspan4 knockout cell line by PCR. e, The average number of migrasomes per 100 µm RF in wild type and Tspan4-KO NRK cells. Data shown represent mean±SEM, n=100 cells for each group. ***P<0.001. f, Statistical analysis of migrasome numbers in wild type L929 and Tspan4-KO L929 cell lines. Data shown represent mean±SEM; n=173 cells for WT L929; n=168 cells for Tspan4-KO L929. NS= No significance. Unprocessed blots related to b were presented in Supplementary Fig. 6. Experiments related to a-f were independently repeated 3 times. The quantifications in e and f are pooled from these independent experiments. Two-tailed unpaired t-test method was used for statistical analysis. The statistical details for a, c, e and f were available in Supplementary Table 4.
Supplementary Figure 2 Dynamics of Tspan4 on migrasomes and RFs.
a, Tspan4-GFP-expressing NRK cells were subjected to FRAP (fluorescent recovery after photo-bleaching) analysis. Migrasomes in the growth phase and the steady phase were imaged by confocal microscopy. Scale bar, 2 µm. b, Area changes of growth-phase and steady-phase migrasomes in (a) during FRAP analysis. c, The normalized mean fluorescence intensity of Tspan4-GFP on growth-phase and steady-phase migrasomes in (a) during FRAP assays. d, Time-lapse images of a RF from a Tspan4-GFP-expressing NRK cell obtained in the course of a FRAP assay using resonant scanning mode confocal microscopy. Scale bar, 2 µm. e, Time-lapse images of a RF taken by resonant scanning mode confocal microscopy of Tspan4-mMaple3 photoconverted from green to red by 405 laser irradiation. Scale bar, 2 µm. Experiments related to a-c were independently repeated 26 times. The quantifications in b and c are one representative experiment. Experiments related to d were independently repeated 5 times. Experiments related to e were independently repeated 8 times.
Supplementary Figure 3 Cholesterol is necessary for migrasome formation.
a, Tspan1-mCherry-, Tspan7-mCherry-, Tspan9-mCherry- and CD82-mCherry-expressing NRK cells were observed by confocal microscopy. Enrichment of Tspan proteins on migrasomes is revealed by heat map images. Scale bars, 10 µm. b, NRK cells stably expressing Tspan4-GFP were treated by MβCD (Methyl-β-cyclodextrin, 10 mM) for 0 min and 30 min. Confocal images were collected at each time point. Scale bar, 10 µm. c, Statistical analysis of migrasome numbers at each time point in images like those in (b). Data shown represent mean±SEM; n=84 cells for MβCD 0 min; n=109 cells for MβCD 30 min. ***P<0.001. d, Relative total cellular cholesterol level of cells grown under different cholesterol depletion conditions, including 10% full cholesterol medium (FBS), cholesterol depletion medium (LPDS) and cholesterol depletion medium with 30 μM Pravastatin. Data shown represent mean±SEM (n=3 independent experiments), ***P<0.001. Experiments related to a-d were independently repeated 3 times. The quantifications in c are pooled from three independent experiments. Two-tailed unpaired t-test method was used for statistical analysis. The statistical details for c and d were available in Supplementary Table 4.
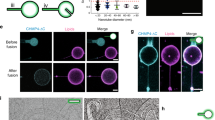
Supplementary Figure 4 Cholesterol is colocalized with Tspan4 on migrasome-like structures in in vitro systems.
Z-stack images of an in vitro-reconstituted Tspan4-containing GUV labeled with Topfluor-cholesterol and stained by Cy5-SE to label Tspan4. Images were acquired by confocal microscopy, then mean intensity Z-projections were generated by ImageJ. Purple, proteins; green, Topfluor-cholesterol; red, Rhodamine-PE. Images are representative of 34 tethers from 4 independent experiments. Scale bar, 2.5 µm.
Supplementary Figure 5 Tspan4 is highly recruited to migrasome-like structures during micropipette-induced reconstitution.
a, GUVs embedded with CD63-GFP were subjected to the in vitro reconstitution assay as described in Fig. 5a. Green, proteins, red, Rhodamine-PE. Images are representative of 10 tethers from 3 independent experiments. Scale bar, 5 µm. b, Time-lapse images showing pulling-induced assembly of tetraspanin-enriched microdomains and formation of a migrasome-like structure by micropipette-induced reconstitution. 6 independent experiments were repeated. Scale bar, 10 µm; zoomed-in panels, 5 µm. c, Working model to explain the analysis method for time-lapse images of micropipette reconstitution using the Imaris Spots function. Tspan4-GFP fluorescent signals on migrasome and retraction fiber structures during micropipette reconstitution were recognized and labeled by Imaris Spots. The initial attachment point of the migrasome or retraction fiber structure was set as the origin. The distance between each spot to the origin was measured and recorded. d, Time-lapse images from (b) were analyzed using the Imaris Spots function. Tspan4-GFP signals labeled by Imaris Spots were traced in each frame and the distance from each spot to the normalized origin was calculated and plotted. 3 independent experiments were repeated. The snapshot merged image of frame 36 is shown in the lower panel; scale bar, 20 µm.
Supplementary Figure 6 Unprocessed Blots and Gel.
a, Unprocessed blots related to Fig. 1d. Verification of Tspan4-GFP protein expression levels in different cell lines by western blot. The sections presented in Fig. 1d were labeled by red squares. b, Unprocessed gel related to Fig. 4a. Recombinant Tspan4-GFP was analyzed on an SDS-PAGE gel and stained by Coomassie Brilliant Blue. The section presented in Fig. 4a was labeled by red square. c, Unprocessed blots related to Supplementary Fig. 1b. Cells with stable Tspan4-GFP expression were treated with or without deglycosylation mix, then lysed and analyzed by western blot. The sections presented in Supplementary Fig. 1b were labeled by red squares.
Supplementary Information
Supplementary Information
Supplementary Figures 1–6, Supplementary Note 1 and legends for Supplementary Tables and Videos.
Supplementary Table 1
Transcription levels of the top 10 expressed TSPAN genes in NRK cells.
Supplementary Table 2
Measurement of liposome radius.
Supplementary Table 3
Measurement of the K1 value of liposomes.
Supplementary Table 4
Statistics source data.
Supplementary Video 1
Tspan4-GFP forms fast-moving puncta on migrasome and retraction fibre. Related to Fig. 2i.
Supplementary Video 2
In vitro reconstitution of migrasome formation by pulling, control GUV. Related to Fig. 5b.
Supplementary Video 3
In vitro reconstitution of migrasome formation by pulling, Tspan4-GFP embedded GUV. Related to Fig. 5b.
Rights and permissions
About this article
Cite this article
Huang, Y., Zucker, B., Zhang, S. et al. Migrasome formation is mediated by assembly of micron-scale tetraspanin macrodomains. Nat Cell Biol 21, 991–1002 (2019). https://doi.org/10.1038/s41556-019-0367-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0367-5
This article is cited by
-
Migrasomal autophagosomes relieve endoplasmic reticulum stress in glioblastoma cells
BMC Biology (2024)
-
Canonical and non-canonical integrin-based adhesions dynamically interconvert
Nature Communications (2024)
-
Mesenchymal stromal cell-associated migrasomes: a new source of chemoattractant for cells of hematopoietic origin
Cell Communication and Signaling (2023)
-
Migrasomes, a new mode of intercellular communication
Cell Communication and Signaling (2023)
-
The versatile roles of testrapanins in cancer from intracellular signaling to cell–cell communication: cell membrane proteins without ligands
Cell & Bioscience (2023)