Abstract
In vertebrates, multipotent progenitors located in the pharyngeal mesoderm form cardiomyocytes and branchiomeric head muscles, but the dynamic gene expression programmes and mechanisms underlying cardiopharyngeal multipotency and heart versus head muscle fate choices remain elusive. Here, we used single-cell genomics in the simple chordate model Ciona to reconstruct developmental trajectories forming first and second heart lineages and pharyngeal muscle precursors and characterize the molecular underpinnings of cardiopharyngeal fate choices. We show that FGF–MAPK signalling maintains multipotency and promotes the pharyngeal muscle fate, whereas signal termination permits the deployment of a pan-cardiac programme, shared by the first and second heart lineages, to define heart identity. In the second heart lineage, a Tbx1/10-Dach pathway actively suppresses the first heart lineage programme, conditioning later cell diversity in the beating heart. Finally, cross-species comparisons between Ciona and the mouse evoke the deep evolutionary origins of cardiopharyngeal networks in chordates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE99846. Previously published microarray data that were reanalysed here are available under accession codes GSE54746. Source data for figures are provided in Supplementary Table 6. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
The code/Rmarkdown files for the analyses reported in this paper are available at https://github.com/ChristiaenLab/single-cell-ciona.
References
Pinto, A. R. et al. Revisiting cardiac cellular composition. Circ. Res. 118, 400–409 (2016).
Saga, Y. et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126, 3437–3447 (1999).
Lescroart, F. et al. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol. 16, 829–840 (2014).
Devine, W. P., . & Wythe, J. D. & George, M. & Koshiba-Takeuchi, K. & Bruneau, B. G. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife 3, e03848 (2014).
Meilhac, S. M., Esner, M., Kelly, R. G., Nicolas, J.-F. & Buckingham, M. E. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell 6, 685–698 (2004).
Kelly, R. G., Brown, N. A. & Buckingham, M. E. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell 1, 435–440 (2001).
Mosimann, C. et al. Chamber identity programs drive early functional partitioning of the heart. Nat. Commun. 6, 8146 (2015).
Nevis, K. et al. Tbx1 is required for second heart field proliferation in zebrafish. Dev. Dyn. 242, 550–559 (2013).
Lescroart, F. et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 359, 1177–1181 (2018).
Diogo, R. et al. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature 520, 466–473 (2015).
Nathan, E. et al. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development 135, 647–657 (2008).
Harel, I. et al. Pharyngeal mesoderm regulatory network controls cardiac and head muscle morphogenesis. Proc. Natl Acad. Sci. USA 109, 18839–18844 (2012).
Tirosh-Finkel, L., Elhanany, H., Rinon, A. & Tzahor, E. Mesoderm progenitor cells of common origin contribute to the head musculature and the cardiac outflow tract. Development 133, 1943–1953 (2006).
Lescroart, F. et al. Clonal analysis reveals common lineage relationships between head muscles and second heart field derivatives in the mouse embryo. Development 137, 3269–3279 (2010).
Gopalakrishnan, S. et al. A cranial mesoderm origin for esophagus striated muscles. Dev. Cell 34, 694–704 (2015).
Mandal, A., Holowiecki, A., Song, Y. C. & Waxman, J. S. Wnt signaling balances specification of the cardiac and pharyngeal muscle fields. Mech. Dev. 143, 32–41 (2017).
Kaplan, N., Razy-Krajka, F. & Christiaen, L. Regulation and evolution of cardiopharyngeal cell identity and behavior: insights from simple chordates. Curr. Opin. Genet. Dev. 32, 119–128 (2015).
Wang, W., Razy-Krajka, F., Siu, E., Ketcham, A. & Christiaen, L. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLoS Biol. 11, e1001725 (2013).
Razy-Krajka, F. et al. Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Dev. Cell 29, 263–276 (2014).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Stolfi, A. et al. Early chordate origins of the vertebrate second heart field. Science 329, 565–568 (2010).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Razy-Krajka, F. et al. An FGF-driven feed-forward circuit patterns the cardiopharyngeal mesoderm in space and time. eLife 7, e29656 (2018).
Grimm, E. C. CONISS: a FORTRAN 77 program for stratigraphically constrained cluster analysis by the method of incremental sum of squares. Comput. Geosci. 13, 13–35 (1987).
Nimmo, R. A., May, G. E. & Enver, T. Primed and ready: understanding lineage commitment through single cell analysis. Trends Cell Biol. 25, 459–467 (2015).
Tolkin, T. & Christiaen, L. Rewiring of an ancestral Tbx1/10-Ebf-Mrf network for pharyngeal muscle specification in distinct embryonic lineages. Development 143, 3852–3862 (2016).
Moris, N., Pina, C. & Arias, A. M. Transition states and cell fate decisions in epigenetic landscapes. Nat. Rev. Genet. 17, 693–703 (2016).
Zhang, L. et al. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev. Biol. 390, 68–79 (2014).
Chen, L., Fulcoli, F. G., Tang, S. & Baldini, A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ. Res. 105, 842–851 (2009).
Liao, J. et al. Identification of downstream genetic pathways of Tbx1 in the second heart field. Dev. Biol. 316, 524–537 (2008).
Davis, R. J., Shen, W., Heanue, T. A. & Mardon, G. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev. Genes Evol. 209, 526–536 (1999).
Kumar, J. P. The molecular circuitry governing retinal determination. Biochim. Biophys. Acta 1789, 306–314 (2009).
Guo, C. et al. A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J. Clin. Invest. 121, 1585–1595 (2011).
Zhou, Z. et al. Temporally distinct Six2-positive second heart field progenitors regulate mammalian heart development and disease. Cell Rep. 18, 1019–1032 (2017).
Stolfi, A., Gandhi, S., Salek, F. & Christiaen, L. Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development 141, 4115–4120 (2014).
Gandhi, S., Haeussler, M., Razy-Krajka, F., Christiaen, L. & Stolfi, A. Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona. Dev. Biol. 425, 8–20 (2017).
Kelly, R. G., Jerome-Majewska, L. A. & Papaioannou, V. E. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum. Mol. Genet. 13, 2829–2840 (2004).
Kong, P. et al. Tbx1 is required autonomously for cell survival and fate in the pharyngeal core mesoderm to form the muscles of mastication. Hum. Mol. Genet. 23, 4215–4231 (2014).
Anderson, H. E. & Christiaen, L. Ciona as a simple chordate model for heart development and regeneration. J. Cardiovasc. Dev. Dis. 3, 25 (2016).
Scialdone, A. et al. Resolving early mesoderm diversification through single-cell expression profiling. Nature 535, 289–293 (2016).
Ibarra-Soria, X. et al. Defining murine organogenesis at single-cell resolution reveals a role for the leukotriene pathway in regulating blood progenitor formation. Nat. Cell Biol. 20, 127–134 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Abu-Issa, R., Smyth, G., Smoak, I., Yamamura, K.-I. & Meyers, E. N. Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development 129, 4613–4625 (2002).
Barron, M., Gao, M. & Lough, J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev. Dyn. 218, 383–393 (2000).
Reifers, F., Walsh, E. C., Léger, S., Stainier, D. Y. & Brand, M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar). Development 127, 225–235 (2000).
Tirosh-Finkel, L. et al. BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development 137, 2989–3000 (2010).
Hutson, M. R. et al. Arterial pole progenitors interpret opposing FGF/BMP signals to proliferate or differentiate. Development 137, 3001–3011 (2010).
Marques, S. R., Lee, Y., Poss, K. D. & Yelon, D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev. Biol. 321, 397–406 (2008).
van Wijk, B. et al. Epicardium and myocardium separate from a common precursor pool by crosstalk between bone morphogenetic protein- and fibroblast growth factor-signaling pathways. Circ. Res. 105, 431–441 (2009).
Zhang, J. et al. Frs2alpha-deficiency in cardiac progenitors disrupts a subset of FGF signals required for outflow tract morphogenesis. Development 135, 3611–3622 (2008).
Vitelli, F., Morishima, M., Taddei, I., Lindsay, E. A. & Baldini, A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum. Mol. Genet. 11, 915–922 (2002).
Zhang, Z., Huynh, T. & Baldini, A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development 133, 3587–3595 (2006).
Li, X. et al. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature 498, 456–462 (2013).
Hotta, K. et al. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 236, 1790–1805 (2007).
Christiaen, L., Wagner, E., Shi, W. & Levine, M. The sea squirt Ciona intestinalis. Cold Spring Harb. Protoc. 2009, db.emo138 (2009).
Wang, W., Racioppi, C., Gravez, B. & Christiaen, L. Purification of fluorescent labeled cells from dissociated Ciona embryos. Adv. Exp. Med. Biol. 1029, 101–107 (2018). in.
Beh, J., Shi, W., Levine, M., Davidson, B. & Christiaen, L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development 134, 3297–3305 (2007).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Chung, N. C. & Storey, J. D. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31, 545–554 (2015).
Maaten, Lvander & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Coifman, R. R. & Lafon, S. Diffusion maps. Appl. Comput. Harmon. Anal. 21, 5–30 (2006).
Haghverdi, L., Buettner, F. & Theis, F. J. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 31, 2989–2998 (2015).
Hastie, T. & Stuetzle, W. Principal curves. J. Am. Stat. Assoc. 84, 502–516 (1989).
van der Maaten, L. Barnes-Hut-SNE. Preprint at https://arxiv.org/abs/1301.3342 (2013).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucl. Acids Res. 41, D377–D386 (2013).
Acknowledgements
We are grateful to F. Razy-Krajka for discussions and sharing reagents before publication. We thank A. Powers for help in processing the single-cell samples in the early phase of this study, and C. Hafemeister and A. Butler for discussion and help with computational analyses. This project was funded by NIH/NHLBI R01 award HL108643 to L.C., trans-Atlantic network of excellence award 15CVD01 from the Leducq Foundation to R.G.K. and L.C. and an NIH New Innovator Award (DP2-HG-009623) to R.S.
Author information
Authors and Affiliations
Contributions
W.W. performed the Ciona experiments. E.J. performed the mouse experiments. X.N., T.S., W.W. and R.S. performed computational analyses. W.M.M., W.W. and R.S. performed the single-cell RNA-seq experiments. W.W., X.N., R.G.K., R.S. and L.C. designed the experiments and analyses. W.W., X.N., R.S. and L.C. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
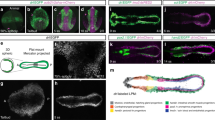
Supplementary Figure 1 Cell clustering and novel cell-type-specific markers.
(a) Violin plots represent the number of genes detected by scRNA-seq in samples obtained from larvae dissociated at different time points (12hpf, 27 cells; 14hpf, 275 cells; 16hpf, 114 cells; 18hpf, 144 cells; 20hpf, 288 cells). The white bars in the centre represent the interquartile range. The black lines in the middle of the white bar represent the median value. The thin black lines extended from the white bar indicate the upper (max) and lower (min) adjacent values in the data. (b) Violin plots represent the distribution of the expression of known outer Atrial Siphon Muscle Precursors (oASMP) markers in 20hpf scRNA-seq dataset (288 cells) as per Razy-Krajka et al.1. (c) t-SNE plots represent clusters of individual transcriptomes from 14, 16, and 18hpf samples (left), and violin plots showing expression of indicated marker genes in defined clusters. (t-SNE is not shown for the 12 hpf data because it consists of a homogeneous population of TVCs). colour codes for cell identities as in Fig. 1a. (a–c) Sample size of the violin plots and t-SNE plots (number of cells in each cell identity) is provided in Supplementary Table 6 (Data sheet: cell identity and number). (d) Figure exemplifying the gating strategy. Cell population only have tagRFP signal in the gate “RFP” were analysed again with defined gates set for tagRFP and tagBFP, cell population with both tagRFP and tagBFP in the gate “RFPBFP” are sorted for downstream genome-wide analysis. Gating strategy is described in detail in method.
Supplementary Figure 2 FISH validation for predicted pan-cardiac markers.
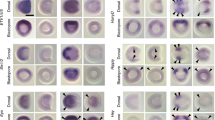
(a) Violin plots and FISH validation of the expression for indicated predicted pan-cardiac genes. (b) Violin plots and FISH validation of the expression for indicated early de novo pan-cardiac markers genes in st.24-25 (14hpf) embryos. (a-b) Violin plots represent the distribution of the expression of indicated genes in defined cell clusters. Sample size of the violin plots (number of cells in each cell identity) is provided in Supplementary Table 6 (Data sheet: cell identity and number). mRNAs are visualized by whole mount fluorescent in situ hybridization (green). Nuclei of TVCs are marked by Mesp>nls::LacZ and revealed by anti-beta-galactosidase antibody (red). Mesp-driven hCD4::mCherry accumulates in the cell membrane and was revealed by anti-mCherry antibody (blue). Anterior to the left. Scale bar, 10 μm. Solid arrowheads, ASM; open arrowheads, SHPs; arrows, FHPs; M, midline (dotted line). The numbers of embryos showing the demonstrated gene expression pattern and numbers of the observed embryos are indicated at the right bottom corner of each image.
Supplementary Figure 3 FISH validation for predicted FHP and SHP-specific markers.
(a) Violin plots and FISH validation of the expression for indicated predicted FHP and SHP-specific genes. Violin plots represent the distribution of the expression of indicated genes in defined cell clusters. Sample size of the violin plots (number of cells in each cell identity) is provided in Supplementary Table 6 (Data sheet: cell identity and number). mRNAs are visualized by whole mount fluorescent in situ hybridization (green). Nuclei of TVCs are marked by Mesp>nls::LacZ and revealed by anti-beta-galactosidase antibody (red). Mesp-driven hCD4::mCherry accumulates in the cell membrane and was revealed by anti-mCherry antibody (blue). Anterior to the left. Scale bar, 10 μm. Solid arrowheads, ASM; open arrowheads, SHPs; arrows, FHPs; M, midline (dotted line). The numbers of embryos showing the demonstrated gene expression pattern and numbers of the observed embryos are indicated at the right bottom corner of each image. (b) Dach and Tbx1/10 regulates the SHP fate specification in the juvenile heart (Unprocessed raw images of Fig. 7c). Dach and Tbx1/10 is required to limit the proportion of SHP-derived cells forming Mhc2+ cardiomyocytes in juveniles. Representative image of analysed embryos. ControlCRISPR, 4 embryos; DachCRISPR, 8 embryos; Tbx1/10CRISPR, 6 embryos. Grey: Mhc2 mRNA visualized by in situ hybridization. SHP-derived cells are labelled with 3XT12>H2B::mCherry (green), B7.5 lineage cells are labelled with Mesp>nls::LacZ (red). Scale bar, 10µm.
Supplementary Figure 4 Pseudotemporal expression profiles of indicated known marker genes along the ASM, FHP and SHP trajectories.
(a) Colour-coded cell identities as defined by unsupervised clustering from larvae dissociated at different time points (12, 14, 16, 18, 20 hpf) (see Fig. 1b, Supplementary Fig. 1c). DC: Diffusion Coordinate. By pooling all the Ciona cardiopharyngeal cells together, the diffusion map successfully captured the split trajectories of pharyngeal vs. cardiac fates, but failed to recover the branching points between the first and second heart lineages. Sample size (number of cells in each cell identity) is provided in Supplementary Table 6 (Data sheet: cell identity and number). (b) Gap statistics plots used to determine the number of transition states in each trajectory. Centre value showed gap statistic with red bar indicating standard error (SE) for 100 bootstraps. Sample size. ASM: 340 cells, FHP: 379 cells, SHP: 247 cells. (c) Pseudotemporal expression profiles of indicated known marker genes along the ASM, FHP and SHP trajectories (Sample size as in b). Y-axes: expression in log2(FPKM). The purple or black lines indicate the smoothed expression profiles. Y-axis: log2(FPKM). X-axis, normalized pseudotime along the principal curve (shown in Fig. 2b). Colours codes as in (a). (d) Raw scaled expression heatmap of temporal expression pattern of indicated cell-type-specific marker genes on ASM, FHP and SHP trajectory. (e) Heatmap of smoothed temporal expression pattern of indicated cell-type specific marker genes on ASM, FHP and SHP trajectory corresponding to (d). Expressions are grouped by k-means clustering.
Supplementary Figure 5 Transcriptional regulation of ASM and FHP fate specification.
(a-b) Histograms with density lines showing predicted induction time of de novo ASM genes or inactivation pseudotime of primed cardiac genes individually with corresponding ASM regulatory states (separated by black dashed lines). (c) Violin plots showing the log fold changes of pan-cardiac genes (n=108) corresponding to Ebf perturbation. The white bars indicate the interquartile range. The black whiskers extended from the bars represent the upper (max) and lower (min) adjacent values in the data. The black lines in the middle of the bars show the median values. (d) Pseudotemporal expression plots showing the progressive depletion of the indicated primed pan-cardiac genes along the ASM trajectory. (e) Pseudotemporal expression profiles of indicated genes along the FHP trajectory. (d-e) X-axis: normalized pseudotime as defined in (b), Y-axes: expression in log2 FPKM. Black lines indicate the smoothed expression. Black dashed lines indicate the transitions between predicted regulatory states, indicated and colour-coded below, and purple dashed lines indicate calculated activation or inactivation pseudotime. Dot colours refer to the sample of origin. (f-g) Density plots showing the number of de novo pan-cardiac genes with calculated activation pseudotime and primed ASM genes with predicted inactivation pseudotime in binned pseudotime windows along FHP trajectories. The black dashed lines indicate the regulatory states. Black dashed lines indicate the transitions between predicted regulatory states and purple dashed lines indicate calculated activation or inactivation pseudotime. (h) Violin plots showing the log fold changes of ASM genes in indicated conditions and time points relative to LacZ controls. Experiment performed in n=2 biological replicates. (i) Heatmap showing log2(fold change) of early FHP and STVC genes (identified in the 14hpf scRNA-seq data) in indicated conditions and time points relative to LacZ controls. (j) Violin plots showing log fold changes for defined categories of genes (FHP and STVC markers identified in the 14hpf scRNA-seq data) in indicated conditions and time points relative to LacZ controls. Experiment performed in n=2 biological replicates. (h-i) The white bars indicate the interquartile range. The black whiskers extended from the bars represent the upper (max) and lower (min) adjacent values in the data. The black lines in the middle of the bars show the median values. Sample size: ASM genes, n= 159; FHP genes, n =18; STVC genes, n= 22.
Supplementary Figure 6 Gene set enrichment statistics of bulk RNA-seq data.
(a-h) Bar plots representing the gene set enrichment statistics reported in Supplementary Table 6 (data sheet: Fig. 5b, S6). In all panels, only ASM and pan-cardiac markers (defined by scRNA-seq, as in Fig. 1) are shown but all expressed genes (log2(CPM)>3) were considered for the statistics. Responses to indicated perturbations of FGF-MAPK signalling at defined time points were categorized into “upregulated”, “downregulated” or “no change” using EdgeR summary statistics based on biological duplicate experiments, using arbitrary cutoffs [|log2(fold change)| > 1 and P-value <0.1]. For each panel, plots on the left show the expected and observed numbers of genes in indicated categories, plots on the right show enrichment as log2(observed/expected). Asterisks indicate significant enrichment, as determined by hypergeometric tests: ***, p < 0.001; **, p<0.01; *, p<0.05. Detailed reports are available in Supplementary Table 6 (data sheet: Fig. 5b, S6). (a-d) At late times points (cardiopharyngeal-lineage cells sorted from 18 or 20 hpf larvae), inhibition of FGF-MAPK signalling upregulated more pan-cardiac markers and downregulated more ASM markers than expected by chance, whereas gain of FGF-MAPK function downregulated more pan-cardiac markers and upregulated more ASM markers than expected by chance. These results are consistent with the hypothesis that FGF-MAPK promotes the ASM programme while inhibiting the pan-cardiac programme.(e-h) At earlier time points, when multipotent cardiopharyngeal progenitors are present in the control samples, loss of FGF-MAPK function downregulated more ASM markers than expected by chance, which is consistent with the notion that expression of primed ASM markers already depends on FGF-MAPK signalling in multipotent progenitors. By contrast, loss of FGF-MAPK function upregulated more of the de novo expressed (and uncertain) pan-cardiac markers than expected by chance, which is consistent with our hypothesis that FGF-MAPK activity in multipotent progenitors prevents the activation of de novo expressed pan-cardiac markers. N numbers in panels a-h represent genes in defined category and are stated for each group in Supplementary Table 6 (data sheet: Fig. 5b, S6).
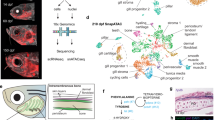
Supplementary Figure 7 Dach1 expression in murine cardiopharyngeal mesoderm and canonical correlation (CC) vector of genes separated cardiac and pharyngeal mesoderm cells in Ciona and Mouse.
(a) Expression patterns of Dach1, Islet1 and Nkx2.5 proteins in E7.5 embryos as indicated. Representative image for two embryos. Bottom panels are close-up views of the regions boxed with a dotted line. Arrows: head folds, Open arrowheads: double Dach1+, Islet1+ pharyngeal mesoderm cell, solid arrowhead: Nkx2.5 expression in a Dach1+, Islet1+ cell. Scale bar, 100 μm. (b) Dach1, Isl1 and Nkx2-5 protein expression detected by immunofluorescence in E8.5 mouse embryos at distinct levels in the pharyngeal mesoderm and developing heart. Representative image for three embryos. Note the broad Dach1 expression, which overlaps with Isl1 in second heart field cells in the dorsal pericardial wall. Scale bar, 100 μm. (c-d) Expression heatmaps of the top 30 genes of canonical correlation vector that contributed most to the heart vs. pharyngeal mesoderm separation in Ciona and the mouse. Detailed information is provided in Supplementary Table 4. (c) Top 30 genes identified from the canonical correlation analysis performed on Ciona and Mouse E6.50- E7.25 single-cell RNA-seq data from Scialdone et al.2. (d) Top 30 genes identified from the canonical correlation analysis performed on Ciona and Mouse Mesp+ E6.75- E7.50 single-cell RNA-seq data from Lescroart et al.3.
Supplementary Figure 8 A conserved cardiopharyngeal programme shared between Ciona and Mouse.
(a) t-SNE plots of Ciona and Mouse E6.50- E7.25 single-cell RNA-seq data from Scialdone et al.2 (Mouse + Ciona cells, n=1163) computed by the top 30 genes of canonical correlation vector that contributed most to the separation of cardiopharyngeal identities in both species. The scaled and centred expression values for these 30 genes were then used to computed a 2-dimension t-SNE embedding for each species separately. Cell classifications are as determined in the original studies of Ciona and Mouse. Box plot showing the classified contribution to each clusters in Ciona and Mouse. t-SNE plots of of Ciona and Mouse scRNA-seq data as described, with the expression patterns of Ebf1 and Gata4. (b) t-SNE plots of Ciona and Mouse Mesp+ E6.75- E7.50 single-cell RNA-seq data from Lescroart et al.3 (Mouse + Ciona cells, n=1361) computed by the top 30 genes of canonical correlation vector that contributed most to the separation of cardiopharyngeal identities in both species. The scaled and centred expression values for these 30 genes were then used to compute a 2-dimension t-SNE embedding for each species separately. Cell classifications are as determined in the original studies of Ciona and Mouse. Box plot showing the classified contribution to each clusters in Ciona and Mouse. t-SNE plots of of Ciona and Mouse scRNA-seq data as described, with the expression patterns of Ebf1 and Gata4. (c) Gene ontology enrichment analysis of conserved cardiopharyngeal markers. Top 30 CC genes were used for each enrichment test, and a set of 11125 orthologs as the background set. Fisher’s exact test was used with the false discovery rate correction for multiple testing. The top CC gene set is significantly enriched in genes encoding transcription factors and DNA binding proteins.
Supplementary information
Supplementary Information
Supplementary Figures 1–8 and titles/legends for Supplementary Tables 1–6 and Supplementary Videos 1 and 2
Supplementary Table 1
Developmental stages in Ciona.
Supplementary Table 2
Top predicted differentially expressed marker genes across different cardiopharyngeal cell types.
Supplementary Table 3
Validation of cell-type-specific gene expression by FISH assays for selected candidate cardiopharyngeal marker genes.
Supplementary Table 4
Canonical correlation vector of genes that separate cardiac and pharyngeal mesoderm cells in Ciona and mouse.
Supplementary Table 5
Ciona-to-mouse gene name dictionary.
Supplementary Table 6
Statistics source data.
Supplementary Video 1
Animated three-dimensional projection of St. 38 juvenile heart.
Supplementary Video 2
Three-dimensional animation of Mesp>LacZ reporter labelled cells in St.38 juvenile.
Rights and permissions
About this article
Cite this article
Wang, W., Niu, X., Stuart, T. et al. A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nat Cell Biol 21, 674–686 (2019). https://doi.org/10.1038/s41556-019-0336-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0336-z
This article is cited by
-
DNA methylation patterns at birth predict health outcomes in young adults born very low birthweight
Clinical Epigenetics (2023)
-
Hand2 delineates mesothelium progenitors and is reactivated in mesothelioma
Nature Communications (2022)
-
Single-cell analysis of cell fate bifurcation in the chordate Ciona
BMC Biology (2021)
-
Cardiopharyngeal deconstruction and ancestral tunicate sessility
Nature (2021)
-
QKI is a critical pre-mRNA alternative splicing regulator of cardiac myofibrillogenesis and contractile function
Nature Communications (2021)