Abstract
Chromatin assembled with the histone H3 variant CENP-A is the heritable epigenetic determinant of human centromere identity. Using genome-wide mapping and reference models for 23 human centromeres, CENP-A binding sites are identified within the megabase-long, repetitive α-satellite DNAs at each centromere. CENP-A is shown in early G1 to be assembled into nucleosomes within each centromere and onto 11,390 transcriptionally active sites on the chromosome arms. DNA replication is demonstrated to remove ectopically loaded, non-centromeric CENP-A. In contrast, tethering of centromeric CENP-A to the sites of DNA replication through the constitutive centromere associated network (CCAN) is shown to enable precise reloading of centromere-bound CENP-A onto the same DNA sequences as in its initial prereplication loading. Thus, DNA replication acts as an error correction mechanism for maintaining centromere identity through its removal of non-centromeric CENP-A coupled with CCAN-mediated retention and precise reloading of centromeric CENP-A.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
ChIP-seq and Repli-seq datasets generated during the current study were deposited at GEO under primary accession number GSE111381. Figures 1, 2, 3, 4, 5 and 6 and Supplementary Figs. 1, 2, 3 and 5 have associated raw mapping data. Mass spectrometry data were deposited at the ProteomeXchange database under accession number PXD013385. Mass spectrometry data are found in Table 1 and Supplementary Fig. 5.
Previously published ChIP-seq data that were reanalysed in this study include HT1080b (ref. 44) CENP-AFLAG ChIP-seq (GSM2808187 and corresponding input dataset GSM2808190) and HuRef (ref. 45) CENP-A native ChIP-seq (GSM1494428, GSM1494429, GSM1494430, GSM1494431, and corresponding input datasets GSM1494424, GSM1494425, GSM1494426, GSM1494427). Hela CENP-ATAP, Hela CENP-ALAP, HT1080b CENP-AFLAG (ref. 44) and HuRef (ref. 45) endogenous CENP-A ChIP-seq datasets were intersected with the following publicly available ENCODE HeLa S3 datasets: DNase I (GSM736564 and GSM736510), H3K4me1 (GSM798322), H3K4me2 (GSM733734), H3K4me3 (GSM733682), H2A.z (GSM1003483), H3K9me3 (GSM1003480), H3K27ac (GSM733684), H3K27me3 (GSM945208), H3K36me3 (GSM733711), H3K79me2 (GSM733669), H3K9ac (GSM733756) and H4K20me1 (GSM733689). CENP-ATAP and CENP-ALAP SICER peaks were also intersected with ENCODE CTCF datasets (GSM749729 and GSM749739), and high-occupancy target region datasets (GSE54296).
Source data for Figs. 1, 3, 5, 6 and 7 and Supplementary Figs. 3, 4 and 5 have been provided as Supplementary Table 4. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
References
Wevrick, R. & Willard, H. F. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proc. Natl Acad. Sci. USA 86, 9394–9398 (1989).
Cleveland, D. W., Mao, Y. & Sullivan, K. F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 (2003).
Willard, H. F. Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 37, 524–532 (1985).
Manuelidis, L. & Wu, J. C. Homology between human and simian repeated DNA. Nature 276, 92–94 (1978).
Earnshaw, W. C. & Rothfield, N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313–321 (1985).
Palmer, D. K., O’Day, K., Wener, M. H., Andrews, B. S. & Margolis, R. L. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104, 805–815 (1987).
Bodor, D. L. et al. The quantitative architecture of centromeric chromatin. eLife 3, e02137 (2014).
Nechemia-Arbely, Y. et al. Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J. Cell Biol. 216, 607–621 (2017).
Sullivan, B. A. & Karpen, G. H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11, 1076–1083 (2004).
Stimpson, K. M. & Sullivan, B. A. Epigenomics of centromere assembly and function. Curr. Opin. Cell Biol. 22, 772–780 (2010).
Marshall, O. J., Chueh, A. C., Wong, L. H. & Choo, K. H. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82, 261–282 (2008).
Fachinetti, D. et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066 (2013).
Foltz, D. R. et al. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 (2006).
Okada, M. et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8, 446–457 (2006).
Hori, T. et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039–1052 (2008).
Hori, T., Shang, W. H., Takeuchi, K. & Fukagawa, T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200, 45–60 (2013).
Padeganeh, A. et al. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr. Biol. 23, 764–769 (2013).
Jansen, L. E., Black, B. E., Foltz, D. R. & Cleveland, D. W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 (2007).
Nechemia-Arbely, Y., Fachinetti, D. & Cleveland, D. W. Replicating centromeric chromatin: spatial and temporal control of CENP-A assembly. Exp. Cell Res. 318, 1353–1360 (2012).
Foltz, D. R. et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009).
Dunleavy, E. M. et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009).
Silva, M. C. et al. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 22, 52–63 (2012).
Lacoste, N. et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 53, 631–644 (2014).
Van Hooser, A. A. et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114, 3529–3542 (2001).
Shrestha, R. L. et al. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget 8, 46781–46800 (2017).
Filipescu, D. et al. Essential role for centromeric factors following p53 loss and oncogenic transformation. Genes Dev. 31, 463–480 (2017).
Mata, J. F., Lopes, T., Gardner, R. & Jansen, L. E. A rapid FACS-based strategy to isolate human gene knockin and knockout clones. PloS ONE 7, e32646 (2012).
Landry, J. J. et al. The genomic and transcriptomic landscape of a HeLa cell line. G3 3, 1213–1224 (2013).
Hayden, K. E. et al. Sequences associated with centromere competency in the human genome. Mol. Cell. Biol. 33, 763–772 (2013).
Conde e Silva, N. et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 370, 555–573 (2007).
Miga, K. H. et al. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24, 697–707 (2014).
Schneider, V. A. et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 27, 849–864 (2017).
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5, e254 (2007).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Zang, C. et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 (2009).
Maloney, K. A. et al. Functional epialleles at an endogenous human centromere. Proc. Natl Acad. Sci. USA 109, 13704–13709 (2012).
Ly, P. et al. Selective Y centromere inactivation triggers chromosome shattering in micronuclei and repair by non-homologous end joining. Nat. Cell Biol. 19, 68–75 (2017).
Amor, D. J. & Choo, K. H. Neocentromeres: role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71, 695–714 (2002).
Hasson, D. et al. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20, 687–695 (2013).
Amor, D. J. et al. Human centromere repositioning “in progress”. Proc. Natl Acad. Sci. USA 101, 6542–6547 (2004).
Alonso, A. et al. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 8, R148 (2007).
Li, H. et al. Functional annotation of HOT regions in the human genome: implications for human disease and cancer. Sci. Rep. 5, 11633 (2015).
Teytelman, L., Thurtle, D. M., Rine, J. & van Oudenaarden, A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl Acad. Sci. USA 110, 18602–18607 (2013).
Thakur, J. & Henikoff, S. Unexpected conformational variations of the human centromeric chromatin complex. Genes Dev. 32, 20–25 (2018).
Henikoff, J. G., Thakur, J., Kasinathan, S. & Henikoff, S. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci. Adv. 1, e1400234 (2015).
Hansen, R. S. et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA 107, 139–144 (2010).
Bui, M. et al. Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. Cell 150, 317–326 (2012).
Erliandri, I. et al. Replication of alpha-satellite DNA arrays in endogenous human centromeric regions and in human artificial chromosome. Nucl. Acids Res. 42, 11502–11516 (2014).
Guse, A., Carroll, C. W., Moree, B., Fuller, C. J. & Straight, A. F. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature 477, 354–358 (2011).
Carroll, C. W., Milks, K. J. & Straight, A. F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143–1155 (2010).
Petrovic, A. et al. Structure of the MIS12 complex and molecular basis of its interaction with CENP-C at human kinetochores. Cell 167, 1028–1040 (2016).
Rago, F., Gascoigne, K. E. & Cheeseman, I. M. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25, 671–677 (2015).
Klare, K. et al. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22 (2015).
Weir, J. R. et al. Insights from biochemical reconstitution into the architecture of human kinetochores. Nature 537, 249–253 (2016).
Hoffmann, S. et al. CENP-A is dispensable for mitotic centromere function after initial centromere/kinetochore assembly. Cell Rep. 17, 2394–2404 (2016).
Smith, S. & Stillman, B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 (1991).
Verreault, A., Kaufman, P. D., Kobayashi, R. & Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87, 95–104 (1996).
Hayashi, T. et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729 (2004).
Kaufman, P. D., Kobayashi, R., Kessler, N. & Stillman, B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81, 1105–1114 (1995).
Huang, H. et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat. Struct. Mol. Biol. 22, 618–626 (2015).
Zhang, W. et al. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 7, 12619 (2016).
Sun, X. et al. Elevated expression of the centromere protein-A (CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int. J. Cancer 139, 899–907 (2016).
Gascoigne, K. E. et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145, 410–422 (2011).
Zasadzinska, E. et al. Inheritance of CENP-A nucleosomes during DNA replication requires HJURP. Dev. Cell 47, 348–362 e7 (2018).
Zhang, J., Kobert, K., Flouri, T. & Stamatakis, A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucl. Acids Res. 32, D493–D496 (2004).
Wang, Z., Wu, C., Aslanian, A., Yates, J. R.III. & Hunter, T. Defective RNA polymerase III is negatively regulated by the SUMO-Ubiquitin-Cdc48 pathway. eLife 7, e35447 (2018).
Acknowledgements
We thank A. Desai, P. Ly and C. Eissler for critical discussion, L.E.T. Jansen (Gulbenkian Institute, Portugal) for reagents and D.-H. Kim for productive discussions and technical help. This work was supported by grants (R01 GM-074150 and R35 GM-122476) from the NIH to D.W.C., who receives salary support from Ludwig Cancer Research.
Author information
Authors and Affiliations
Contributions
Y.N.-A. and D.W.C. conceived and designed experiments and wrote the manuscript. Y.N.-A. performed experiments. K.H.M. analysed the sequencing data. M.A.M. and O.S. analysed data and performed experiments. D.F. suggested experiments and provided key experimental input. A.Y.L. and B.R. prepared sequencing libraries and provided resources. A.A. and J.R.Y. performed mass spectrometry experiments and provided resources.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Identification of peaks enriched for CENP-A binding.
(a) Scheme showing experimental design for tagging an endogenous CENP-A locus to produce CENP-A+/LAP HeLa cells. These cells were then adapted to suspension growth. (b) Scheme showing the experimental design for obtaining increased levels of CENP-ATAP expression. CENP-ATAP is expressed in these cells at 4.5-fold the level of CENP-A in the parental HeLa cells1. (c, d) Localization of endogenously tagged CENP-ALAP (c) and CENP-ATAP (d) determined with indirect immunofluorescence using anti-GFP antibody (c) or rabbit IgG (d). Scale bar, 5 μm. The experiment was repeated independently three times with similar results for both (c) and (d). (e) FACS analysis of DNA content showing the synchronization efficiency of CENP-A+/LAP and CENP-ATAP HeLa cell lines. The experiment was repeated independently five times with similar results. (f, g) Examples of centromeric regions of chromosome 7 (f) and 5 (g) showing increased occupancy of overexpressed CENP-ATAP (compare CENP-ATAP with CENP-ALAP). The experiment was repeated independently twice with similar results. (h) Overlap between G1 and G2 CENP-ATAP binding peaks at α-satellite sequences. (i) Top, overlap between G1 and G2 CENP-A single-mapping binding sites at α-satellite HOR sequences. Bottom, peak overlap between G1 CENP-ATAP (increased expression) and CENP-ALAP (endogenous level) single-mapping binding sites at α-satellite HOR sequences.
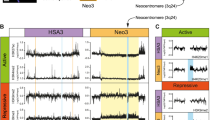
Supplementary Figure 2 CENP-A ChIP-seq identifies CENP-A binding at reference centromeres of 23 human chromosomes.
CENP-ALAP bound DNAs at G1 and G2 were sequenced, with 2 replicates per condition, and mapped to the centromeric reference models in the hg38 assembly2,3. Shown are the raw mapping data (coloured) for every human centromere (except for the centromere of chromosome 19 that shares almost all of its α-satellites arrays with α-satellites arrays of chromosomes 1 and 5) and CENP-A binding called as SICER peaks (black lines, underneath) for one replicate for each time point. The experiment was repeated twice independently with similar results. Centromere reference location, red. CENP-B box, orange.
Supplementary Figure 3 Ectopic deposition of CENP-A into open and active chromatin at G1 does not function as a seeding hotspot for neocentromere formation.
(a) Number of non-α-satellite CENP-A SICER binding sites called at G1 or G2 at different fold thresholds (above background). (b) Human DLD1 cells with auxin degradable CENP-AAID and a doxycycline-inducible CENP-AWT4, were synchronized at G1 using the CDK4/6 inhibitor PD-0332991 (also known as Palbociclib) or at mitosis using nocodazole, following addition of doxycycline. The experiment was repeated independently three times with similar results. (c) Read mapping data of CENP-ATAP ChIP-seq at G1 (red) and G2 (blue), at the chromosomal location of a known patient derived neocentromere5 found in chromosome 13. The experiment was repeated twice independently with similar results. A third human neocentromere, identified in line MS4221, has been identified to lie within a 400 kb neocentromere at position 86.5 to 86.9 Mb on chromosome 8 in hg195,6 (corresponding to 85.78–85.88 Mb in hg38). However, a gap and segmental duplications that appear in this region precluded precise analysis of CENP-A mapping at this neocentromere. (d-g) Fold enrichment of CENP-ATAP chromatin in randomly cycling cells or at G1 (d, e) and CENP-ALAP chromatin (f, g) at G1 at different genomic locations. SICER peaks ≥ 5-fold supported between two replicates were analysed for their enrichment level at different genomic locations, compared to the level of enrichment at these sites by chance. (h, i) Number of CENP-ATAP (h) and CENP-ALAP (i) SICER peaks ≥ 5-fold that overlap with ‘HOT’ regions in the human genome in G1 and G2 synchronized cells. Data shown are from two biologically independent experiments. Source data for d-i can be found in Supplementary Table 4.
Supplementary Figure 4 Non-centromeric CENP-A binding peaks overlap with active transcription marks.
(a,b) The chromatin features of CENP-ATAP (a) and CENP-ALAP (b) non-centromeric preferential sites were analysed by intersecting SICER peaks ≥ 5-fold supported between two replicates with publicly available ENCODE datasets for histone modification profiles in HeLa S3, that represent modifications typically associated with transcription activation or repression. The experiment was performed one time, except for DNase I and H3K27me3 for which there are 2 ENCODE datasets available, and therefore for DNase I and H3K27me3 the experiment was repeated twice independently with similar results. Statistics source data for Supplementary Fig. 4a, b can be found in Supplementary Table 4. (The sum of ectopic CENP-ATAP sites at active or repression marks is more than 100%, the result of overlap between H3K9me3 and active transcription marks.) (c,d) The chromatin features of sites of preferential, non-centromeric CENP-A binding were analysed for histone modification profiles associated with transcription activation or repression in HeLa S3 cells by intersecting SICER peaks ≥ 5-fold found in previously published CENP-A ChIP-seq datasets in HT10807 (c) and HuRef8 (d) cell lines with publicly available ENCODE datasets for histone modification profiles in HeLa S3. For HT1080b (c) the experiment was performed one time, except for DNase I and H3K27me3 for which there are 2 ENCODE datasets available, and therefore for DNase I and H3K27me3 the experiment was repeated twice independently with similar results. For HuRef (d), The experiment was performed four times, except for DNase I and H3K27me3 for which there are 2 ENCODE datasets available, and therefore for DNase I and H3K27me3 the experiment was repeated independently eight times with similar results. Source data for a-d can be found in Supplementary Table 4.
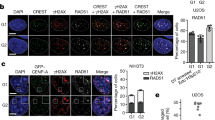
Supplementary Figure 5 Centromeres are late replicating with CENP-A remaining tethered locally by continued binding to the CCAN complex.
(a) FACS analysis of DNA content showing the synchronization efficiency of CENP-ATAP HeLa cell line across S phase. The experiment was repeated independently twice with similar results. (b) Genomic DNA of cells labelled for 1 hour with BrdU was sonicated prior to the BrdU immunoprecipitation and fragments of 200–800 bp were obtained. The experiment was repeated independently twice with similar results. Unprocessed images of DNA gels can be found in Supplementary Fig. 6. (c) Quantitative real-time PCR for MRGPRE and MMP15 genes, previously reported to replicate early (ref9 and ENCODE Repli-seq). (d) Quantitative real-time PCR for HBE1 and Sat210 genes, previously reported to replicate late (ref11 and ENCODE Repli-seq). (e) Quantitative real-time PCR for α-satellite DNA. Data shown in c-e are from two biologically independent experiments. Source data for c-e can be found in Supplementary Table 4. (f) MNase digestion profile showing the nucleosomal DNA length distributions of bulk input mononucleosomes (upper panel) and purified CENP-ATAP following native ChIP at early S and mid-S phase. The experiment was repeated twice independently with similar results. (g) CENP-A ChIP-seq raw mapping data spanning the whole of cen18 at G1, mid-S phase and G2, and BrdU repli-seq at early S (S1), mid-S (S4) and late S/G2 (S7). SICER peaks are denoted as black lines underneath the raw mapping data. The experiment was repeated twice independently with similar results. Centromere reference location, red. CENP-B boxes, orange. Scale bar, 2Mb. (h) Overlap degree between CENP-A G1 and mid-S at α-satellite HORs single copy variants. (i) Ethidium Bromide stained DNA agarose gel showing MNase digestion profile of bulk chromatin used for mass spectrometry identification of proteins associating with CENP-ATAP chromatin (left panel) and for CENP-ATAP co-immunoprecipitation experiment (right panel). Mass spectrometry was performed once and co-IP was performed twice with similar results. Unprocessed images of DNA gels can be found in Supplementary Fig. 6. (j-n) CENP-ATAP immunopurification followed by mass spectrometry identifies association with CENP-A chromatin of DNA replication related proteins (j,k), chromatin remodelling factors and nuclear chaperones (l), histones (m) and centromere and kinetochore proteins (n).
Supplementary Figure 6
Unprocessed film scans of all immunoblots and DNA gels with corresponding protein and DNA size markers.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and their legends, and legends for Supplementary Video 1 and Supplementary Tables 1–4
Supplementary Table 1
Read statistics for ChIP-seq and Repli-seq experiments.
Supplementary Table 2
Endogenous CENP-A sequence mapping onto α-satellite DNAs in human centromere reference models for each autosome and the X chromosome.
Supplementary Table 3
Antibodies used in the study.
Supplementary Table 4
Statistical source data for graphical representations.
Supplementary Video 1
Rapid CENP-CAE/AE depletion following IAA treatment.
Rights and permissions
About this article
Cite this article
Nechemia-Arbely, Y., Miga, K.H., Shoshani, O. et al. DNA replication acts as an error correction mechanism to maintain centromere identity by restricting CENP-A to centromeres. Nat Cell Biol 21, 743–754 (2019). https://doi.org/10.1038/s41556-019-0331-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0331-4
This article is cited by
-
Higher-order protein assembly controls kinetochore formation
Nature Cell Biology (2024)
-
DNAJC9 prevents CENP-A mislocalization and chromosomal instability by maintaining the fidelity of histone supply chains
The EMBO Journal (2024)
-
Stable inheritance of H3.3-containing nucleosomes during mitotic cell divisions
Nature Communications (2022)
-
CENPA promotes clear cell renal cell carcinoma progression and metastasis via Wnt/β-catenin signaling pathway
Journal of Translational Medicine (2021)
-
CENP-A overexpression promotes distinct fates in human cells, depending on p53 status
Communications Biology (2021)