Abstract
Cellular senescence is a stable growth arrest that is implicated in tissue ageing and cancer. Senescent cells are characterized by an upregulation of proinflammatory cytokines, which is termed the senescence-associated secretory phenotype (SASP). NAD+ metabolism influences both tissue ageing and cancer. However, the role of NAD+ metabolism in regulating the SASP is poorly understood. Here, we show that nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme of the NAD+ salvage pathway, governs the proinflammatory SASP independent of senescence-associated growth arrest. NAMPT expression is regulated by high mobility group A (HMGA) proteins during senescence. The HMGA–NAMPT–NAD+ signalling axis promotes the proinflammatory SASP by enhancing glycolysis and mitochondrial respiration. HMGA proteins and NAMPT promote the proinflammatory SASP through NAD+-mediated suppression of AMPK kinase, which suppresses the p53-mediated inhibition of p38 MAPK to enhance NF-κB activity. We conclude that NAD+ metabolism governs the proinflammatory SASP. Given the tumour-promoting effects of the proinflammatory SASP, our results suggest that anti-ageing dietary NAD+ augmentation should be administered with precision.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Code availability
The software and algorithms for data analyses used in this study are all well established from previous work and are referenced throughout the manuscript. No custom code was used in this study.
Data availability
ChIP-seq data have been deposited into the GEO under accession number GSE111841. Raw RNA-seq data for RAS-induced senescence and control conditions in IMR90 cells were downloaded from GEO (under accession code GSE74324)41. For the correlation analysis between NAMPT and SASP genes in human laser capture and microdissected PanIN lesion samples, gene expression data were obtained from GEO (under accession code GSE43288)1. Source data used for statistical analyses of Figs. 1a,b,e,h,i, 2b,c,e,f,h,j, 3a–n, 4a,c,e,f,h,j,l,m,p, 5a,d–g, 6b–e,g–i, 7b,c,e,f,g, and Supplementary Figs. 2c–f,h–j,l–n,p,q, 3a–f,h, 4, 5a–d, 6b–g,i,k,l, and 7a,b,d,e are provided as Supplementary Table 1 (Statistics source data). All other data supporting the findings of this study are available upon request.
References
Hayflick, L. & Moorhead, P. S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961).
Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013).
Wang, X. et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 49, 289–295 (2017).
Childs, B. G. et al. Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov. 16, 718–735 (2017).
Sieben, C. J., Sturmlechner, I., van de Sluis, B. & van Deursen, J. M. Two-step senescence-focused cancer therapies. Trends Cell Biol. 28, 723–737 (2018).
Hoare, M. et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat. Cell Biol. 18, 979–992 (2016).
Ito, Y., Hoare, M. & Narita, M. Spatial and temporal control of senescence. Trends Cell Biol. 27, 820–832 (2017).
Sumter, T. F. et al. The high mobility group A1 (HMGA1) transcriptome in cancer and development. Curr. Mol. Med. 16, 353–393 (2016).
Bianchi, M. E. & Agresti, A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opini. Genet. Dev. 15, 496–506 (2005).
Thomas, J. O. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 29, 395–401 (2001).
Narita, M. et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126, 503–514 (2006).
Verdin, E. NAD(+) in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015).
Garten, A. et al. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat. Rev. Endocrinol. 11, 535–546 (2015).
Martin, P. R., Shea, R. J. & Mulks, M. H. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 183, 1168–1174 (2001).
Rongvaux, A. et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32, 3225–3234 (2002).
Wiley, C. D. et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314 (2016).
Zhang, H. et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 (2016).
Mills, K. F. et al. Long-term administration of nicotinamide mononucleotide mitigates age-associated physiological decline in mice. Cell Metab. 24, 795–806 (2016).
Hasmann, M. & Schemainda, I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 63, 7436–7442 (2003).
Hara, N., Yamada, K., Shibata, T., Osago, H. & Tsuchiya, M. Nicotinamide phosphoribosyltransferase/visfatin does not catalyze nicotinamide mononucleotide formation in blood plasma. PLoS ONE 6, e22781 (2011).
Rielland, M. et al. Senescence-associated SIN3B promotes inflammation and pancreatic cancer progression. J. Clin. Invest. 124, 2125–2135 (2014).
Guerra, C. et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 19, 728–739 (2011).
Nelson, D. L. & Cox, M. M. Lehninger Principles of Biochemistry 6th Edn (W. H. Freeman, New York, 2013).
Hardie, D. G., Ross, F. A. & Hawley, S. A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262 (2012).
Zhou, G. et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174 (2001).
Jones, R. G. et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18, 283–293 (2005).
Coppe, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Berger, F., Lau, C., Dahlmann, M. & Ziegler, M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 280, 36334–36341 (2005).
Freund, A., Patil, C. K. & Campisi, J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 30, 1536–1548 (2011).
van der Veer, E. et al. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J. Biol. Chem. 282, 10841–10845 (2007).
Lim, C. S., Potts, M. & Helm, R. F. Nicotinamide extends the replicative life span of primary human cells. Mech. Ageing Dev. 127, 511–514 (2006).
Di Micco, R. et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat. Cell Biol. 13, 292–302 (2011).
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006).
Shah, S. N. & Resar, L. M. High mobility group A1 and cancer: potential biomarker and therapeutic target. Histol. Histopathol. 27, 567–579 (2012).
Rodier, F. et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009).
Davalos, A. R., Coppe, J. P., Campisi, J. & Desprez, P. Y. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 29, 273–283 (2010).
Yoshimoto, S. et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013).
Zhang, R. et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 (2005).
Liu, P. et al. Regulation of inflammatory cytokine expression in pulmonary epithelial cells by pre-B-cell colony-enhancing factor via a nonenzymatic and AP-1-dependent mechanism. J. Biol. Chem. 284, 27344–27351 (2009).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Tasdemir, N. et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 6, 612–629 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, (15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).
Crnogorac-Jurcevic, T. et al. Molecular analysis of precursor lesions in familial pancreatic cancer. PLoS ONE 8, e54830 (2013).
Acknowledgements
The authors thank the Wistar Institute Proteomics and Metabolomics Facility for technical assistance. This work was supported by grants from the following organizations: the US National Institutes of Health (R01CA160331, R01CA163377, R01CA202919 and P50CA228991 to R.Z.; P01AG031862 to R.Z., K.N. and D.S.; R01CA148639 and R21CA155736 to G.D.; F31CA206387 to L.L.; R00CA194309 to K.M.A.; R01DK098656 to J.A.B.; R01CA131582 to D.W.S.; R50CA211199 to A.V.K.; R50CA221838 to H.-Y.T.; and T32CA009191 to T.N.); the US Department of Defense (OC140632P1 and OC150446 to R.Z.); The Honorable Tina Brozman Foundation for Ovarian Cancer Research (to R.Z.); and the Ovarian Cancer Research Alliance (Collaborative Research Development Grant to R.Z. and D.W.S., and Ann and Sol Schreiber Mentored Investigator Award to S.W.). Support of Core Facilities was provided by Cancer Centre Support Grant CA010815 to The Wistar Institute.
Author information
Authors and Affiliations
Contributions
T.N., L.L., T.F., J.Z., N.F., S.W., K.M.A., O.I., A.V.K., D.S., Z.S. and H.-Y.T. performed the experiments and analysed data. T.N., G.D. and R.Z. designed the experiments. J.A.B. and D.W.S. contributed to the study design. K.N., G.D. and R.Z. supervised studies. R.Z. conceived the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
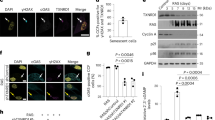
Supplementary Figure 1 NAMPT is a direct HMGA1 target gene.
a, List of putative direct HMGA1 target genes identified by cross-referencing HMGA1 ChIP-seq dataset with RNA-seq dataset during oncogenic-RAS induced senescence in IMR90 cells. b, HMGA1 ChIP-seq tracks for control and oncogenic RAS-induced senescent cells at the human NAMPT gene locus. c, Schematic of experimental design and reference time frame unless otherwise stated.
Supplementary Figure 2 HMGA1-mediated NAMPT expression regulates senescence.
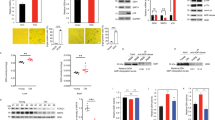
a-d Cells were induced to senesce by oncogenic RAS with or without the indicated shRNAs or treated with 1nM FK866 at senescent initiation. Expression of the indicated proteins was determined by immunoblot (a) or cells were subjected to colony formation and SA-β-Gal staining (b). Integrated intensity of the colonies formed by the indicated cells (c) or the percentage of SA-β-gal positive cells was quantified (d). e, Cells were induced to senesce by oncogenic RAS and analyzed at the indicated time points for expression of TGFβ1 and TGFβ3 mRNA by qRT-PCR. f, Cells were induced to senesce by oncogenic RAS with or without the indicated shRNAs or treated with 1 nM FK866 at senescence initiation. Expression of the indicated SASP genes was determined by qRT-PCR. g-j, In established senescent cells, HMGA1 or NAMPT was knocked down using the indicated shRNAs. NAMPT activity was also inhibited by 1 nM FK866. The indicated cells were subjected to colony formation or SA-β-gal staining (g). SA-β-gal activity (h) and integrated intensity of the colonies formed by the indicated cells were quantified (i). j-m, In established senescent cells induced by oncogenic BRAF, HMGA1 or NAMPT were knocked down using the indicated shRNAs. NAMPT activity was also inhibited by 1 nM FK866. Expression of NAMPT and the indicated SASP genes were determined by qRT-PCR (j). Cells were subjected colony formation or SA-β-gal staining (k). SA-β-gal activity (l) and integrated intensity of the colonies formed by the indicated cells were quantified (m). n-q, In established etoposide-induced senescent cells, HMGA1 or NAMPT were knocked down using the indicated shRNAs. NAMPT activity was also inhibited by 1 nM FK866. Expression of NAMPT and SASP mRNA were determined using qRT-PCR (n). Cells were subjected colony formation or SA-β-gal staining (o). SA-β-gal activity (p) and integrated intensity of the colonies formed by the indicated cells were quantified (q). r, In established senescent cells, HMGA1 or NAMPT were knocked down using the indicated shRNAs. NAMPT activity was also inhibited by 1 nM FK866. Cells were cultured in serum-free media for 48 hours. Total protein was extracted from the media and run on an immunoblot to detect NAMPT and coomassie stained to show equal loading. Scale bar = 100 μm. n=3 independent experiments for all panels. All graphs represent mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are provided in Supplementary Table 1. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 8.
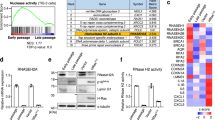
Supplementary Figure 3 Oncogene and therapy-induced senescence display increased NAD+/NADH ratio.
a,b, In established RAS-induced senescent cells, HMGA1 or NAMPT was knocked down using the indicated shRNAs. NAMPT activity was also inhibited by FK866. Steady-state NAD+ metabolite levels were measured by LC-MS/MS (a), or determined using the NAD+/NADH-Glo Assay. (b). c,d, In established senescent cells induced by oncogenic BRAFV600E (c) or etoposide (50 μM for 48 hours) (d), HMGA1 or NAMPT was knocked down using the indicated shRNAs. NAMPT activity was also inhibited by 1 nM FK866. The NAD+/NADH ratio was measured using the NAD+/NADH-Glo Assay. e, In established senescent cells induced by oncogenic RAS, HMGA1 or NAMPT was knocked down using the indicated shRNAs. NAMPT activity was also inhibited by 1 nM FK866. Expression of IL10 and TNFα mRNA were measured using qRT-PCR. f, Cells with or without ectopic V5-tagged HMGA1 expression with or without NAMPT knockdown were examined for the NAD+ levels using the NAD+/NADH-Glo Assay. g, The gating strategy used for determining mean fluorescence intensity. h, In established senescent cells induced by oncogenic RAS, HMGA1 or NAMPT was knocked down using the indicated shRNAs. NAMPT activity was also inhibited by 1 nM FK866. Cells were analyzed using the glycolysis stress test on a Seahorse Bioanalyzer XFe96. Representative glycolysis stress test profile displaying the ECAR in response to the sequential injection of glucose (10 mM), oligomycin A (2 μM), and 2-deoxyglucose (50 mM) (number of independent experiments is indicated). n=3 independent experiments for all panels unless otherwise stated on figures. All graphs represent mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are provided in Supplementary Table 1.
Supplementary Figure 4 NAMPT expression correlates with expression of SASP genes in PanIN lesions.
Pearson correlation analysis of NAMPT with the indicated SASP genes in human laser capture and microdissected (LCM) PanIN lesion samples based on the GSE43288 dataset. n=13 biologically independent samples. P values were calculated using a Pearson correlation analysis. Statistical source data are provided in Supplementary Table 1.
Supplementary Figure 5 NAD+ metabolism drives the proinflammatory SASP.
a, b, In established senescent cells induced by RAS (a) or etoposide (b), cells were treated with gallotannin. Expression of the indicated SASP genes was measured using qRT-PCR. c,d, In established senescent cells, HMGA1 (c) or NAMPT (d) was knocked down. NAMPT activity was also inhibited by 1 nM FK866 (d). Expression of CEBPβ mRNA was measured using qRT-PCR. n=3 independent experiments for all panels. All graphs represent mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are provided in Supplementary Table 1.
Supplementary Figure 6 HMGA1/NAMPT-regulated high proinflammatory SASP is absent in replicative senescence.
a-c, NAMPT activity was inhibited by knocking down using the indicated shNAMPTs or treated with 1 nM FK866. Cells were subjected colony formation or SA-β-gal staining (a). SA-β-gal activity (b) and integrated intensity of the colonies formed by the indicated cells were quantified (c). d-e, Cells cultured at early passage (population doubling 30, PD30), late passage (population doubling 90, PD90), and under oncogene-induced senescence (PD30 expressing oncogenic RAS) were compared for the NAD+/NADH ratio (d) and expression of the indicated proinflammatory SASP genes by qRT-PCR (e). f, The indicated cells with or without NAM supplementation were compared for expression of the indicated SASP genes determined by qRT-PCR. g, The indicated cells with or without NA supplementation were compared for expression of the indicated SASP genes determined by qRT-PCR. h-i, IMR90 cells were induced to undergo senescence by knocking down of NAMPT or HMGA1 or inhibition of NAMPT activity by 1 nM FK866. RAS induced senescent cells were used as a positive control. Expression of the indicated proteins was determined using immunoblot (h). Expression of the indicated SASP genes was determined by qRT-PCR (i). j, IMR90 cells were induced to senesce by oncogenic RAS and analyzed at the indicated time points for expression of the indicated proteins by immunoblot. k-l, Cells from the conditions in (d) were analyzed using a Seahorse Bioanalyzer XFe96 for extracellular acidification (ECAR) (l) and oxygen consumption (OCR) (m). Data were normalized based on protein concentration. Scale bar = 100 μm. n=3 independent experiments for all panels unless otherwise stated on figures. All graphs represent mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are provided in Supplementary Table 1. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 8.
Supplementary Figure 7 Increasing SASP by oncogenic RAS expression or NMN supplementation in replicative senescent cells does not affect senescence-associated growth arrest.
a, The indicated early passage (population doubling 30, PD30), late passage (population doubling 90, PD90), late passage expressing oncogenic RAS (PD90 expressing oncogenic RAS) (a) or supplemented with NMN (b) were compared for NFκb reporter activities. c-e, Cells cultured at early passage (population doubling 30), late passage (population doubling 90), late passage expressing RAS (population doubling 90), and late passage treated with NMN (population doubling 90) were subjected to colony formation or SA-β-gal staining (c). SA-β-gal activity (d) and integrated intensity of the colonies formed by the indicated cells were quantified (e). Scale bar = 100 μm. n=3 independent experiments for all panels. All graphs represent mean ± s.d. P values were calculated using a two-tailed t-test. Statistical source data are provided in Supplementary Table 1.
Supplementary Figure 8 Unprocessed images of all gels and blots.
Unprocessed images of scanned immunoblots shown in Figures and Supplementary Figures are provided.
Supplementary information
Supplementary Information
Supplementary Figures 1–8 and legend for Supplementary Table 1.
Supplementary Table 1
Statistics source data
Rights and permissions
About this article
Cite this article
Nacarelli, T., Lau, L., Fukumoto, T. et al. NAD+ metabolism governs the proinflammatory senescence-associated secretome. Nat Cell Biol 21, 397–407 (2019). https://doi.org/10.1038/s41556-019-0287-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-019-0287-4
This article is cited by
-
Modulation of fracture healing by senescence-associated secretory phenotype (SASP): a narrative review of the current literature
European Journal of Medical Research (2024)
-
Cholesterol biosynthetic pathway induces cellular senescence through ERRα
npj Aging (2024)
-
TXNRD1 drives the innate immune response in senescent cells with implications for age-associated inflammation
Nature Aging (2024)
-
Age-, sex- and proximal–distal-resolved multi-omics identifies regulators of intestinal aging in non-human primates
Nature Aging (2024)
-
Dysregulation of histone modifications in bone marrow mesenchymal stem cells during skeletal ageing: roles and therapeutic prospects
Stem Cell Research & Therapy (2023)