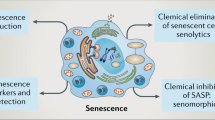
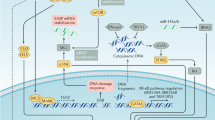
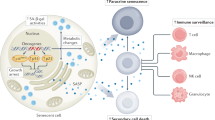
Abstract
Cellular senescence is implicated in physiological and pathological processes spanning development, wound healing, age-related decline in organ functions and cancer. Here, we discuss cell-autonomous and non-cell-autonomous properties of senescence in the context of tumour formation and anticancer therapy, and characterize these properties, such as reprogramming into stemness, tissue remodelling and immune crosstalk, as far more dynamic than suggested by the common view of senescence as an irreversible, static condition.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hayflick, L. & Moorhead, P. S. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961).
Childs, B. G., Durik, M., Baker, D. J. & van Deursen, J. M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015).
Shay, J. W. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 (2016).
Kuilman, T., Michaloglou, C., Mooi, W. J. & Peeper, D. S. The essence of senescence. Genes Dev. 24, 2463–2479 (2010).
Kuilman, T. & Peeper, D. S. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 9, 81–94 (2009).
Coppe, J. P., Desprez, P. Y., Krtolica, A. & Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 (2010).
Perez-Mancera, P. A., Young, A. R. & Narita, M. Inside and out: the activities of senescence in cancer. Nat. Rev. Cancer 14, 547–558 (2014).
Chandra, T. et al. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 10, 471–483 (2015).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic Ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997).
Narita, M. et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 (2003).
Coppe, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Salama, R., Sadaie, M., Hoare, M. & Narita, M. Cellular senescence and its effector programs. Genes Dev. 28, 99–114 (2014).
Sharpless, N. E. & Sherr, C. J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015).
Seshadri, T. & Campisi, J. Repression of c-fos transcription and an altered genetic program in senescent human fibroblasts. Science 247, 205–209 (1990).
Beausejour, C. M. et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 22, 4212–4222 (2003).
Sage, J., Miller, A. L., Perez-Mancera, P. A., Wysocki, J. M. & Jacks, T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature 424, 223–228 (2003).
Milanovic, M. et al. Senescence-associated reprogramming promotes cancer stemness. Nature 553, 96–100 (2018).
Yu, Y. et al. Targeting the senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell 33, 322–336 (2018).
Mosteiro, L. et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 354, aaf4445 (2016).
Chiche, A. et al. Injury-induced senescence enables in vivo reprogramming in skeletal muscle. Cell Stem Cell 20, 407–414.e4 (2017).
Latella, L. et al. DNA damage signaling mediates the functional antagonism between replicative senescence and terminal muscle differentiation. Genes Dev. 31, 648–659 (2017).
Ritschka, B. et al. The senescence-associated secretory phenotype induces cellular plasticity and tissue regeneration. Genes Dev. 31, 172–183 (2017).
Giancotti, F. G. Mechanisms governing metastatic dormancy and reactivation. Cell 155, 750–764 (2013).
Sosa, M. S., Bragado, P. & Aguirre-Ghiso, J. A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer 14, 611–622 (2014).
De Cecco, M. et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12, 247–256 (2013).
Ivanov, A. et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 202, 129–143 (2013).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Kang, C. et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612 (2015).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Gluck, S. et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017).
Salmina, K. et al. Nucleolar aggresomes mediate release of pericentric heterochromatin and nuclear destruction of genotoxically treated cancer cells. Nucleus 8, 205–221 (2017).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA 114, E4612–E4620 (2017).
Di Micco, R. et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 (2006).
Fumagalli, M. et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat. Cell Biol. 14, 355–365 (2012).
Hoare, M. et al. NOTCH1 mediates a switch between two distinct secretomes during senescence. Nat. Cell Biol. 18, 979–992 (2016).
Schmitt, C. A. The persistent dynamic secrets of senescence. Nat. Cell Biol. 18, 913–915 (2016).
Quijano, C. et al. Oncogene-induced senescence results in marked metabolic and bioenergetic alterations. Cell Cycle 11, 1383–1392 (2012).
Dorr, J. R. et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature 501, 421–425 (2013).
Kaplon, J. et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112 (2013).
Kang, T. W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Ohtani, N. et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409, 1067–1070 (2001).
Lin, A. W. et al. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12, 3008–3019 (1998).
Bartkova, J. et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 (2006).
Ferbeyre, G. et al. PML is induced by oncogenic Ras and promotes premature senescence. Genes Dev. 14, 2015–2027 (2000).
Zhang, R. et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1A and HIRA. Dev. Cell 8, 19–30 (2005).
Narita, M. et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126, 503–514 (2006).
Ye, X. et al. Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1A-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2452–2465 (2007).
Chandra, T. et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol. Cell 47, 203–214 (2012).
Sadaie, M. et al. Redistribution of the lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev. 27, 1800–1808 (2013).
Shah, P. P. et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 27, 1787–1799 (2013).
Rai, T. S. et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev. 28, 2712–2725 (2014).
Tasdemir, N. et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 6, 612–629 (2016).
Hewitt, G. et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 3, 708 (2012).
Suram, A. et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J. 31, 2839–2851 (2012).
Rodier, F. et al. DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. J. Cell Sci. 124, 68–81 (2011).
Sulli, G., Di Micco, R. & d’Adda di Fagagna, F. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer 12, 709–720 (2012).
Rapisarda, V. et al. Integrin β3 regulates cellular senescence by activating the TGF-β pathway. Cell Rep. 18, 2480–2493 (2017).
Ito, T., Teo, Y. V., Evans, S. A., Neretti, N. & Sedivy, J. M. Regulation of cellular senescence by Polycomb chromatin modifiers through distinct DNA damage- and histone methylation-dependent pathways. Cell Rep. 22, 3480–3492 (2018).
Acosta, J. C. et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133, 1006–1018 (2008).
Kuilman, T. et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 (2008).
Chien, Y. et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011).
Jing, H. et al. Opposing roles of NF-κB in anti-cancer treatment outcome unveiled by cross-species investigations. Genes Dev. 25, 2137–2146 (2011).
Shimi, T. et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 25, 2579–2593 (2011).
Takahashi, A. et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 8, 15287 (2017).
Ahn, J., Gutman, D., Saijo, S. & Barber, G. N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl Acad. Sci. USA 109, 19386–19391 (2012).
Ablasser, A. et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013).
Xue, W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007).
Chuprin, A. et al. Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes Dev. 27, 2356–2366 (2013).
Biran, A. et al. Senescent cells communicate via intercellular protein transfer. Genes Dev. 29, 791–802 (2015).
Krtolica, A., Parrinello, S., Lockett, S., Desprez, P. Y. & Campisi, J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl Acad. Sci. USA 98, 12072–12077 (2001).
Ohanna, M. et al. Senescent cells develop a PARP-1 and nuclear factor-κB-associated secretome (PNAS). Genes Dev. 25, 1245–1261 (2011).
Lasry, A. & Ben-Neriah, Y. Senescence-associated inflammatory responses: aging and cancer perspectives. Trends Immunol. 36, 217–228 (2015).
Davalos, A. R. et al. p53-dependent release of alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol. 201, 613–629 (2013).
Kortlever, R. M., Higgins, P. J. & Bernards, R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 8, 877–884 (2006).
Acosta, J. C. et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 15, 978–990 (2013).
Eggert, T. et al. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell 30, 533–547 (2016).
Reimann, M. et al. Tumor stroma-derived TGF-β limits Myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell 17, 262–272 (2010).
van Riggelen, J. et al. The interaction between Myc and Miz1 is required to antagonize TGFβ-dependent autocrine signaling during lymphoma formation and maintenance. Genes Dev. 24, 1281–1294 (2010).
Braumuller, H. et al. T-helper-1-cell cytokines drive cancer into senescence. Nature 494, 361–365 (2013).
Wang, L. et al. High-throughput functional genetic and compound screens identify targets for senescence induction in cancer. Cell Rep. 21, 773–783 (2017).
Debacq-Chainiaux, F., Erusalimsky, J. D., Campisi, J. & Toussaint, O. Protocols to detect senescence-associated β-galactosidase (SA-βgal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 (2009).
Bash, J. et al. Rel/NF-κB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors. EMBO J. 18, 2803–2811 (1999).
Schwarzer, R., Dorken, B. & Jundt, F. Notch is an essential upstream regulator of NF-κB and is relevant for survival of Hodgkin and Reed-Sternberg cells. Leukemia 26, 806–813 (2012).
Pribluda, A. et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell 24, 242–256 (2013).
Schwitalla, S. et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 23, 93–106 (2013).
Hernandez-Segura, A. et al. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 27, 2652–2660.e4 (2017).
Toso, A. et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep. 9, 75–89 (2014).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods 14, 873–876 (2017).
Georgilis, A. et al. PTBP1-mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell 34, 85–102.e9 (2018).
Zhu, J. et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654 (2013).
Munoz-Espin, D. et al. Programmed cell senescence during mammalian embryonic development. Cell 155, 1104–1118 (2013).
Storer, M. et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155, 1119–1130 (2013).
Demaria, M. et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014).
Michaloglou, C. et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005).
Haugstetter, A. M. et al. Cellular senescence predicts treatment outcome in metastasised colorectal cancer. Br. J. Cancer 103, 505–509 (2010).
Tamayo-Orrego, L. et al. Evasion of cell senescence leads to medulloblastoma progression. Cell Rep. 14, 2925–2937 (2016).
Banito, A. et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 23, 2134–2139 (2009).
Hong, H. et al. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460, 1132–1135 (2009).
Kawamura, T. et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144 (2009).
Li, H. et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139 (2009).
Marion, R. M. et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460, 1149–1153 (2009).
Utikal, J. et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460, 1145–1148 (2009).
Cruickshanks, H. A. et al. Senescent cells harbour features of the cancer epigenome. Nat. Cell Biol. 15, 1495–1506 (2013).
Waddington, C. H. The Strategy of the Genes: a Discussion of Some Aspects of Theoretical Biology (Allen & Unwin, London, 1957).
Wajapeyee, N., Serra, R. W., Zhu, X., Mahalingam, M. & Green, M. R. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 132, 363–374 (2008).
Neves, J., Demaria, M., Campisi, J. & Jasper, H. Of flies, mice, and men: evolutionarily conserved tissue damage responses and aging. Dev. Cell 32, 9–18 (2015).
Aarts, M. et al. Coupling shRNA screens with single-cell RNA-seq identifies a dual role for mTOR in reprogramming-induced senescence. Genes Dev. 31, 2085–2098 (2017).
Schmitt, C. A. et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109, 335–346 (2002).
Braig, M. et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005).
Collado, M. et al. Tumour biology: senescence in premalignant tumours. Nature 436, 642 (2005).
Lazzerini Denchi, E., Attwooll, C., Pasini, D. & Helin, K. Deregulated E2F activity induces hyperplasia and senescence-like features in the mouse pituitary gland. Mol. Cell. Biol. 25, 2660–2672 (2005).
Baker, D. J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Baker, D. J. et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
Demaria, M. et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 (2017).
Farr, J. N. et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 23, 1072–1079 (2017).
Jeon, O. H. et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 23, 775–781 (2017).
Xu, M. et al. JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proc. Natl Acad. Sci. USA 112, E6301–E6310 (2015).
Zhu, Y. et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658 (2015).
Zhu, Y. et al. New agents that target senescent cells: the flavone, fisetin, and the BCL-XL inhibitors, A1331852 and A1155463. Aging (Albany NY) 9, 955–963 (2017).
Schmitt, C. A. UnSASPing senescence: unmasking tumor suppression? Cancer Cell 34, 6–8 (2018).
Nardella, C., Clohessy, J. G., Alimonti, A. & Pandolfi, P. P. Pro-senescence therapy for cancer treatment. Nat. Rev. Cancer 11, 503–511 (2011).
Kirkland, J. L. & Tchkonia, T. Cellular senescence: a translational perspective. EBioMedicine 21, 21–28 (2017).
Narita, M. et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970 (2011).
Young, A. R. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009).
Lehmann, B. D. et al. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 68, 7864–7871 (2008).
Schafer, M. J. et al. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 8, 14532 (2017).
Acknowledgements
This work was supported by grants to C.A.S. from the Deutsche Forschungsgemeinschaft (SCHM 1633/9-1 and SCHM 1633/11-1), by the Helmholtz Alliance ‘Preclinical Comprehensive Cancer Center’ grant (no. HA-305) from the Helmholtz Association and by the Deutsche Krebshilfe (grant no. 110678). This work was further made possible by the Berlin School of Integrative Oncology (BSIO) graduate program, funded within the German Excellence Initiative and the German Cancer Consortium (GCC), to S.L. and C.A.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lee, S., Schmitt, C.A. The dynamic nature of senescence in cancer. Nat Cell Biol 21, 94–101 (2019). https://doi.org/10.1038/s41556-018-0249-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0249-2
This article is cited by
-
A novel aging-associated lncRNA signature for predicting prognosis in osteosarcoma
Scientific Reports (2024)
-
Blocking methionine catabolism induces senescence and confers vulnerability to GSK3 inhibition in liver cancer
Nature Cancer (2024)
-
Cellular senescence gene TACC3 associated with colorectal cancer risk via genetic and DNA methylated alteration
Archives of Toxicology (2024)
-
A transcriptomic pan-cancer signature for survival prognostication and prediction of immunotherapy response based on endothelial senescence
Journal of Biomedical Science (2023)
-
Single-cell omics traces the heterogeneity of prostate cancer cells and the tumor microenvironment
Cellular & Molecular Biology Letters (2023)