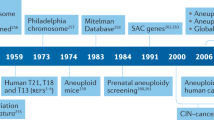
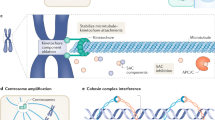
Abstract
Aneuploidy, or imbalanced chromosome number, has profound effects on eukaryotic cells. In humans, aneuploidy is associated with various pathologies, including cancer, which suggests that it mediates a proliferative advantage under these conditions. Here, we discuss physiological changes triggered by aneuploidy, such as altered cell growth, transcriptional changes, proteotoxic stress, genomic instability and response to interferons, and how cancer cells adapt to the changing aneuploid genome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Peter, J. et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018).
Duan, S. F. et al. The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat. Commun. 9, 2690 (2018).
Pilo, D., Carvalho, S., Pereira, P., Gaspar, M. B. & Leitao, A. Is metal contamination responsible for increasing aneuploidy levels in the Manila clam Ruditapes philippinarum? Sci. Total Environ. 577, 340–348 (2017).
Tumova, P., Uzlikova, M., Jurczyk, T. & Nohynkova, E. Constitutive aneuploidy and genomic instability in the single-celled eukaryote Giardia intestinalis. Microbiologyopen 5, 560–574 (2016).
Leitao, A., Boudry, P. & Thiriot-Quievreux, C. Evidence of differential chromosome loss in aneuploid karyotypes of the Pacific oyster. Crassostrea gigas. Genome 44, 735–737 (2001).
Selmecki, A., Forche, A. & Berman, J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313, 367–370 (2006).
Lee, A. J. et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71, 1858–1870 (2011).
Swanton, C. et al. Chromosomal instability determines taxane response. Proc. Natl Acad. Sci. USA 106, 8671–8676 (2009).
Kuznetsova, A. Y. et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle 14, 2810–2820 (2015).
Turajlic, S. & Swanton, C. Metastasis as an evolutionary process. Science 352, 169–175 (2016).
Bakhoum, S. F. et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018).
Heng, H. H. et al. Stochastic cancer progression driven by non-clonal chromosome aberrations. J. Cell Physiol. 208, 461–472 (2006).
McClelland, S. E. Role of chromosomal instability in cancer progression. Endocr. Relat. Cancer 24, T23–T31 (2017).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Hassold, T. & Hunt, P. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 (2001).
Holubcova, Z., Blayney, M., Elder, K. & Schuh, M. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 348, 1143–1147 (2015).
Delhanty, J. D. et al. Detection of aneuploidy and chromosomal mosaicism in human embryos during preimplantation sex determination by fluorescent in situ hybridisation, (FISH). Hum. Mol. Genet. 2, 1183–1185 (1993).
van Echten-Arends, J. et al. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum. Reprod. Update 17, 620–627 (2011).
Bazrgar, M., Gourabi, H., Valojerdi, M. R., Yazdi, P. E. & Baharvand, H. Self-correction of chromosomal abnormalities in human preimplantation embryos and embryonic stem cells. Stem Cells Dev. 22, 2449–2456 (2013).
Bolton, H. et al. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat. Commun. 7, 11165 (2016).
Knouse, K. A., Wu, J., Whittaker, C. A. & Amon, A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc. Natl Acad. Sci. USA 111, 13409–13414 (2014).
Rehen, S. K. et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl Acad. Sci. USA 98, 13361–13366 (2001).
Rehen, S. K. et al. Constitutional aneuploidy in the normal human brain. J. Neurosci. 25, 2176–2180 (2005).
Yurov, Y. B. et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS ONE 2, e558 (2007).
Duncan, A. W. et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology 142, 25–28 (2012).
Schoenfelder, K. P. et al. Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development 141, 3551–3560 (2014).
Weaver, B. A. & Cleveland, D. W. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18, 658–667 (2006).
Storchova, Z. & Kuffer, C. The consequences of tetraploidy and aneuploidy. J. Cell Sci. 121, 3859–3866 (2008).
Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689 (2018).
Gordon, D. J., Resio, B. & Pellman, D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13, 189–203 (2012).
Nagaoka, S. I., Hassold, T. J. & Hunt, P. A. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13, 493–504 (2012).
Levine, M. S. & Holland, A. J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 32, 620–638 (2018).
Cheeseman, I. M. The kinetochore. Cold Spring Harb. Perspect. Biol. 6, a015826 (2014).
Lara-Gonzalez, P., Westhorpe, F. G. & Taylor, S. S. The spindle assembly checkpoint. Curr. Biol. 22, R966–R980 (2012).
Lampson, M. A. & Grishchuk, E. L. Mechanisms to avoid and correct erroneous kinetochore–microtubule attachments. Biology (Basel) 6, 1 (2017).
Sansregret, L. & Swanton, C. The role of aneuploidy in cancer evolution. Cold Spring Harb. Perspect. Med. 7, a028373 (2017).
Simonetti, G., Bruno, S., Padella, A., Tenti, E. & Martinelli, G. Aneuploidy: cancer strength or vulnerability? Int. J. Cancer https://doi.org/10.1002/ijc.31718 (2018).
Hernando, E. et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430, 797–802 (2004).
Manning, A. L., Longworth, M. S. & Dyson, N. J. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 24, 1364–1376 (2010).
Cui, Y., Borysova, M. K., Johnson, J. O. & Guadagno, T. M. Oncogenic B-RafV600E induces spindle abnormalities, supernumerary centrosomes, and aneuploidy in human melanocytic cells. Cancer Res. 70, 675–684 (2010).
Orr, B. & Compton, D. A. A double-edged sword: how oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front. Oncol. 3, 164 (2013).
Conery, A. R. & Harlow, E. High-throughput screens in diploid cells identify factors that contribute to the acquisition of chromosomal instability. Proc. Natl Acad. Sci. USA 107, 15455–15460 (2010).
Meena, J. K. et al. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 34, 1371–1384 (2015).
Knouse, K. A., Lopez, K. E., Bachofner, M. & Amon, A. Chromosome segregation fidelity in epithelia requires tissue architecture. Cell 175, 200–211 (2018).
Castedo, M. et al. Cell death by mitotic catastrophe: a molecular definition. Oncogene 23, 2825–2837 (2004).
Janssen, A., Kops, G. J. & Medema, R. H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl Acad. Sci. USA 106, 19108–19113 (2009).
Thompson, S. L. & Compton, D. A. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188, 369–381 (2010).
Hinchcliffe, E. H. et al. Chromosome missegregation during anaphase triggers p53 cell cycle arrest through histone H3.3 Ser31 phosphorylation. Nat. Cell Biol. 18, 668–675 (2016).
Santaguida, S. et al. Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651 (2017).
Soto, M. et al. p53 prohibits propagation of chromosome segregation errors that produce structural aneuploidies. Cell Rep. 19, 2423–2431 (2017).
Lopez-Garcia, C. et al. BCL9L dysfunction impairs caspase-2 expression permitting aneuploidy tolerance in colorectal cancer. Cancer Cell 31, 79–93 (2017).
Sansregret, L. et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov. 7, 218–233 (2017).
Torres, E. M. et al. Identification of aneuploidy-tolerating mutations. Cell 143, 71–83 (2010).
Donnelly, N., Passerini, V., Durrbaum, M., Stingele, S. & Storchova, Z. HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 33, 2374–2387 (2014).
Dodgson, S. E., Santaguida, S., Kim, S., Sheltzer, J. & Amon, A. The pleiotropic deubiquitinase Ubp3 confers aneuploidy tolerance. Genes Dev. 30, 2259–2271 (2016).
Michel, L. S. et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409, 355–359 (2001).
Sotillo, R. et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11, 9–23 (2007).
Weaver, B. A., Silk, A. D., Montagna, C., Verdier-Pinard, P. & Cleveland, D. W. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36 (2007).
Foijer, F. et al. Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy-induced stress. Proc. Natl Acad. Sci. USA 111, 13427–13432 (2014).
Ohashi, A. et al. Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat. Commun. 6, 7668 (2015).
Fournier, R. E. A general high-efficiency procedure for production of microcell hybrids. Proc. Natl Acad. Sci. USA 78, 6349–6353 (1981).
Upender, M. B. et al. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 64, 6941–6949 (2004).
Stingele, S. et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 8, 608 (2012).
Jiang, J. et al. Translating dosage compensation to trisomy 21. Nature 500, 296–300 (2013).
Adikusuma, F., Williams, N., Grutzner, F., Hughes, J. & Thomas, P. Targeted deletion of an entire chromosome using CRISPR/Cas9. Mol. Ther. 25, 1736–1738 (2017).
Zuo, E. et al. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 18, 224 (2017).
Williams, B. R. et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709 (2008).
Torres, E. M. et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317, 916–924 (2007).
Pavelka, N. et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325 (2010).
Beach, R. R. et al. Aneuploidy causes non-genetic individuality. Cell 169, 229–242 (2017).
Naylor, R. M. & van Deursen, J. M. Aneuploidy in cancer and aging. Annu. Rev. Genet. 50, 45–66 (2016).
Zhu, J., Tsai, H.-J., Gordon, M. R. & Li, R. Cellular stress associated with aneuploidy. Dev. Cell 44, 420–431 (2018).
Segal, D. J. & McCoy, E. E. Studies on Down’s syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. J. Cell Physiol. 83, 85–90 (1974).
Thorburn, R. R. et al. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol. Biol. Cell 24, 1274–1289 (2013).
Ariyoshi, K. et al. Induction of genomic instability and activation of autophagy in artificial human aneuploid cells. Mutat. Res. 790, 19–30 (2016).
Bonney, M. E., Moriya, H. & Amon, A. Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage-sensitive genes. Genes Dev. 29, 898–903 (2015).
Gogendeau, D. et al. Aneuploidy causes premature differentiation of neural and intestinal stem cells. Nat. Commun. 6, 8894 (2015).
Jonas, K., Liu, J., Chien, P. & Laub, M. T. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154, 623–636 (2013).
Barr, A. R. et al. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 8, 14728 (2017).
Liu, X. et al. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev. Dyn. 209, 85–91 (1997).
Ben-David, U. et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat. Commun. 5, 4825 (2014).
Zhang, M. et al. Aneuploid embryonic stem cells exhibit impaired differentiation and increased neoplastic potential. EMBO J. 35, 2285–2300 (2016).
Selmecki, A., Forche, A. & Berman, J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9, 991–1008 (2010).
Yona, A. H. et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl Acad. Sci. USA 109, 21010–21015 (2012).
Rutledge, S. D. et al. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci. Rep. 6, 22828 (2016).
Baker, D. E. et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 25, 207–215 (2007).
Huettel, B., Kreil, D. P., Matzke, M. & Matzke, A. J. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 4, e1000226 (2008).
Mao, R. et al. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol. 6, R107 (2005).
Lockstone, H. E. et al. Gene expression profiling in the adult Down syndrome brain. Genomics 90, 647–660 (2007).
Halevy, T., Biancotti, J. C., Yanuka, O., Golan-Lev, T. & Benvenisty, N. Molecular characterization of Down syndrome embryonic stem cells reveals a role for RUNX1 in neural differentiation. Stem Cell Rep. 7, 777–786 (2016).
Aziz, N. M. et al. Lifespan analysis of brain development, gene expression and behavioral phenotypes in the Ts1Cje, Ts65Dn and Dp(16)1/Yey mouse models of Down syndrome. Dis. Model. Mech. 11, dmm031013 (2018).
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S. & Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996).
Tartaglia, N. R., Howell, S., Sutherland, A., Wilson, R. & Wilson, L. A review of trisomy X (47,XXX). Orphanet J. Rare Dis. 5, 8 (2010).
Stenberg, P. et al. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 5, e1000465 (2009).
Zhang, Y. et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8, e1000320 (2010).
Johansson, A. M., Stenberg, P., Bernhardsson, C. & Larsson, J. Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 26, 2307–2316 (2007).
Gelbart, M. E. & Kuroda, M. I. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136, 1399–1410 (2009).
Hose, J. et al. Dosage compensation can buffer copy-number variation in wild yeast. eLife 4, e05462 (2015).
Torres, E. M., Springer, M. & Amon, A. No current evidence for widespread dosage compensation in S. cerevisiae. eLife 5, e10996 (2016).
Tucker, C. et al. Transcriptional regulation on aneuploid chromosomes in divers Candida albicans mutants. Sci. Rep. 8, 1630 (2018).
Dephoure, N. et al. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3, e03023 (2014).
Liu, Y. et al. Systematic proteome and proteostasis profiling in human trisomy 21 fibroblast cells. Nat. Commun. 8, 1212 (2017).
Vigano, C. et al. Quantitative proteomic and phosphoproteomic comparison of human colon cancer DLD-1 cells differing in ploidy and chromosome stability. Mol. Biol. Cell 29, 1031–1047 (2018).
McShane, E. et al. Kinetic analysis of protein stability reveals age-dependent degradation. Cell 167, 803–815 (2016).
Gasch, A. P. et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000).
Sheltzer, J. M., Torres, E. M., Dunham, M. J. & Amon, A. Transcriptional consequences of aneuploidy. Proc. Natl Acad. Sci. USA 109, 12644–12649 (2012).
O’Duibhir, E. et al. Cell cycle population effects in perturbation studies. Mol. Syst. Biol. 10, 732 (2014).
Sheltzer, J. M. A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res. 73, 6401–6412 (2013).
Durrbaum, M. et al. Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genomics 15, 139 (2014).
Tang, Y. C., Williams, B. R., Siegel, J. J. & Amon, A. Identification of aneuploidy-selective antiproliferation compounds. Cell 144, 499–512 (2011).
Oromendia, A. B., Dodgson, S. E. & Amon, A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 26, 2696–2708 (2012).
Stefani, M. & Dobson, C. M. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J. Mol. Med. (Berl.) 81, 678–699 (2003).
Hanna, J. et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 (2006).
Santaguida, S. & Amon, A. Aneuploidy triggers a TFEB-mediated lysosomal stress response. Autophagy 11, 2383–2384 (2015).
Sheltzer, J. M. et al. Aneuploidy drives genomic instability in yeast. Science 333, 1026–1030 (2011).
Nicholson, J. M. et al. Chromosome mis-segregation and cytokinesis failure in trisomic human cells. eLife 4, e05068 (2015).
Passerini, V. et al. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 7, 10754 (2016).
Sharma, K. et al. Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol. Cell. Proteomics 11, M111.014654 (2012).
Spurgers, K. B. et al. Identification of cell cycle regulatory genes as principal targets of p53-mediated transcriptional repression. J. Biol. Chem. 281, 25134–25142 (2006).
Tan, Y. H., Schneider, E. L., Tischfield, J. & Epstein, C. J. & Ruddle, F. H. Human chromosome 21 dosage: effect on the expression of the interferon induced antiviral state. Science 186, 61–63 (1974).
Iwamoto, T. et al. Influences of interferon-γ on cell proliferation and interleukin-6 production in Down syndrome derived fibroblasts. Arch. Oral Biol. 54, 963–969 (2009).
Sullivan, K. D. et al. Trisomy 21 consistently activates the interferon response. eLife 5, e16220 (2016).
Ling, K. H. et al. Functional transcriptome analysis of the postnatal brain of the Ts1Cje mouse model for Down syndrome reveals global disruption of interferon-related molecular networks. BMC Genomics 15, 624 (2014).
Tanaka, M. H. et al. Expression of interferon-γ, interferon-α and related genes in individuals with Down syndrome and periodontitis. Cytokine 60, 875–881 (2012).
Maroun, L. E. Interferon action and chromosome 21 trisomy (Down syndrome): 15 years later. J. Theor. Biol. 181, 41–46 (1996).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Lan, Y. Y., Londono, D., Bouley, R., Rooney, M. S. & Hacohen, N. Dnase2a deficiency uncovers lysosomal clearance of damaged nuclear DNA via autophagy. Cell Rep. 9, 180–192 (2014).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Bartsch, K. et al. Absence of RNase H2 triggers generation of immunogenic micronuclei removed by autophagy. Hum. Mol. Genet. 26, 3960–3972 (2017).
Gasser, S., Orsulic, S., Brown, E. J. & Raulet, D. H. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436, 1186–1190 (2005).
Rodier, F. et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009).
Cohen, I. et al. IL-1α is a DNA damage sensor linking genotoxic stress signaling to sterile inflammation and innate immunity. Sci. Rep. 5, 14756 (2015).
Andriani, G. A. et al. Whole chromosome instability induces senescence and promotes SASP. Sci. Rep. 6, 35218 (2016).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA 114, E4612–E4620 (2017).
Hardy, P. A. & Zacharias, H. Reappraisal of the Hansemann–Boveri hypothesis on the origin of tumors. Cell Biol. Int. 29, 983–992 (2005).
Knouse, K. A., Davoli, T., Elledge, S. J. & Amon, A. Aneuploidy in cancer: Seq-ing answers to old questions. Annu. Rev. Cancer Biol. 1, 335–354 (2017).
Sheltzer, J. M. et al. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31, 240–255 (2017).
Iwanaga, Y. et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 67, 160–166 (2007).
Dai, W. et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 64, 440–445 (2004).
Baker, D. J. et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36, 744–749 (2004).
Baker, D. J. et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J. Cell Biol. 172, 529–540 (2006).
Hasle, H., Clemmensen, I. H. & Mikkelsen, M. Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet 355, 165–169 (2000).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Dai, C. & Sampson, S. B. HSF1: guardian of proteostasis in cancer. Trends Cell Biol. 26, 17–28 (2016).
Perera, R. M. et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature 524, 361–365 (2015).
Sidera, K. & Patsavoudi, E. HSP90 inhibitors: current development and potential in cancer therapy. Recent Pat. Anticancer Drug Discov. 9, 1–20 (2014).
Bastola, P., Oien, D. B., Cooley, M. & Chien, J. Emerging cancer therapeutic targets in protein homeostasis. AAPS J. 20, 94 (2018).
Mahalingam, D. et al. Targeting HSP90 for cancer therapy. Br. J. Cancer 100, 1523–1529 (2009).
Whitesell, L. & Lindquist, S. L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772 (2005).
Burrell, R. A. et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 494, 492–496 (2013).
Chan, Y. W., Fugger, K. & West, S. C. Unresolved recombination intermediates lead to ultra-fine anaphase bridges, chromosome breaks and aberrations. Nat. Cell Biol. 20, 92–103 (2018).
Duesberg, P., Rausch, C., Rasnick, D. & Hehlmann, R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl Acad. Sci. USA 95, 13692–13697 (1998).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Kops, G. J., Foltz, D. R. & Cleveland, D. W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl Acad. Sci. USA 101, 8699–8704 (2004).
Godek, K. M. et al. Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discov. 6, 532–545 (2016).
Beaupere, C. et al. Genetic screen identifies adaptive aneuploidy as a key mediator of ER stress resistance in yeast. Proc. Natl Acad. Sci. USA 15, 9586–9591 (2018).
Chen, G., Bradford, W. D., Seidel, C. W. & Li, R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482, 246–250 (2012).
Rancati, G. et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135, 879–893 (2008).
Kaya, A. et al. Adaptive aneuploidy protects against thiol peroxidase deficiency by increasing respiration via key mitochondrial proteins. Proc. Natl Acad. Sci. USA 112, 10685–10690 (2015).
Ryu, H.-Y., Wilson, N. R., Mehta, S., Hwang, S. S. & Hochstrasser, M. Loss of the SUMO protease Ulp2 triggers a specific multichromosome aneuploidy. Genes Dev. 30, 1881–1894 (2016).
Millet, C., Ausiannikava, D., Le Bihan, T., Granneman, S. & Makovets, S. Cell populations can use aneuploidy to survive telomerase insufficiency. Nat. Commun. 6, 8664 (2015).
Hughes, T. R. et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25, 333–337 (2000).
Chen, G. et al. Targeting the adaptability of heterogeneous aneuploids. Cell 160, 771–784 (2015).
Vesely, M. D., Kershaw, M. H., Schreiber, R. D. & Smyth, M. J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29, 235–271 (2011).
Dunn, G. P., Bruce, A. T., Ikeda, H., Old, L. J. & Schreiber, R. D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 3, 991–998 (2002).
Schreiber, R. D., Old, L. J. & Smyth, M. J. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570 (2011).
McGranahan, N. et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271 (2017).
Acknowledgements
The work on aneuploidy is supported by the German Research Foundation, grant agreement STO918/5-1 to Z.S.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chunduri, N.K., Storchová, Z. The diverse consequences of aneuploidy. Nat Cell Biol 21, 54–62 (2019). https://doi.org/10.1038/s41556-018-0243-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0243-8
This article is cited by
-
The two sides of chromosomal instability: drivers and brakes in cancer
Signal Transduction and Targeted Therapy (2024)
-
Aneuploid embryonic stem cells drive teratoma metastasis
Nature Communications (2024)
-
Two decades of chromosomal instability and aneuploidy
Chromosome Research (2024)
-
Mutant p53 gains oncogenic functions through a chromosomal instability-induced cytosolic DNA response
Nature Communications (2024)
-
Restricting epigenetic activity promotes the reprogramming of transformed cells to pluripotency in a line-specific manner
Cell Death Discovery (2023)