Abstract
The linear-ubiquitin chain assembly complex (LUBAC) modulates signalling via various immune receptors. In tumour necrosis factor (TNF) signalling, linear (also known as M1) ubiquitin enables full gene activation and prevents cell death. However, the mechanisms underlying cell death prevention remain ill-defined. Here, we show that LUBAC activity enables TBK1 and IKKε recruitment to and activation at the TNF receptor 1 signalling complex (TNFR1-SC). While exerting only limited effects on TNF-induced gene activation, TBK1 and IKKε are essential to prevent TNF-induced cell death. Mechanistically, TBK1 and IKKε phosphorylate the kinase RIPK1 in the TNFR1-SC, thereby preventing RIPK1-dependent cell death. This activity is essential in vivo, as it prevents TNF-induced lethal shock. Strikingly, NEMO (also known as IKKγ), which mostly, but not exclusively, binds the TNFR1-SC via M1 ubiquitin, mediates the recruitment of the adaptors TANK and NAP1 (also known as AZI2). TANK is constitutively associated with both TBK1 and IKKε, while NAP1 is associated with TBK1. We discovered a previously unrecognized cell death checkpoint that is mediated by TBK1 and IKKε, and uncovered an essential survival function for NEMO, whereby it enables the recruitment and activation of these non-canonical IKKs to prevent TNF-induced cell death.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The raw data for RNA-seq analysis (Fig. 2; Supplementary Fig. 2c; Supplementary Table 3) and the proteomic raw data (Figs. 1a and 5f; Supplementary Tables 2, 4 and 5) are available as described below. The RNA-seq raw dataset generated during the current study are available in the SRA repository and can be accessed using the BioProject accession number PRJNA422567 or the SRA accession number SRP126844 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP126844). The proteomic raw data have been deposited in the ProteomeXchange Consortium via the PRIDE70 partner repository with the dataset identifiers PXD008497 (TNFR1-SC analysis), PXD010777 (TBK1 analysis) and PXD008518 (RIPK1 kinase assay). Source data for the graphs of all the other experiments in this study are available in Supplementary Table 1, and unprocessed scans for western blots are displayed in Supplementary Fig. 7. Publicly available tools were used for RNA-seq analysis as specified in the online methods, and the corresponding computational code is available upon request directly with the authors.
References
Micheau, O. & Tschopp, J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 (2003).
Kalliolias, G. D. & Ivashkiv, L. B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 12, 49–62 (2016).
Brenner, D., Blaser, H. & Mak, T. W. Regulation of tumour necrosis factor signalling: live or let die. Nat. Rev. Immunol. 15, 362–374 (2015).
Hrdinka, M. & Gyrd-Hansen, M. The Met1-linked ubiquitin machinery: emerging themes of (de)regulation. Mol. Cell 68, 265–280 (2017).
Shimizu, Y., Taraborrelli, L. & Walczak, H. Linear ubiquitination in immunity. Immunol. Rev. 266, 190–207 (2015).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471, 637–641 (2011).
Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011).
Tokunaga, F. et al. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011).
Peltzer, N. et al. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9, 153–165 (2014).
Emmerich, C. H. et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl Acad. Sci. USA 110, 15247–15252 (2013).
Peltzer, N. et al. LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature 557, 112–117 (2018).
Haas, T. L. et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 (2009).
Draber, P. et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 13, 2258–2272 (2015).
Kupka, S., Reichert, M., Draber, P. & Walczak, H. Formation and removal of poly-ubiquitin chains in the regulation of tumor necrosis factor-induced gene activation and cell death. FEBS J. 283, 2626–2639 (2016).
Rahighi, S. et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136, 1098–1109 (2009).
Dondelinger, Y. et al. NF-kappaB-independent role of IKKalpha/IKKbeta in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol. Cell 60, 63–76 (2015).
Menon, M. B. et al. p38MAPK/MK2-dependent phosphorylation controls cytotoxic RIPK1 signalling in inflammation and infection. Nat. Cell Biol. 19, 1248–1259 (2017).
Jaco, I. et al. MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol. Cell 66, 698–710 (2017).
Dondelinger, Y. et al. MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat. Cell Biol. 19, 1237–1247 (2017).
Kotlyarov, A. et al. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1, 94–97 (1999).
Kanayama, A. et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol. Cell 15, 535–548 (2004).
Wang, C. et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001).
Adhikari, A., Xu, M. & Chen, Z. J. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226 (2007).
Wu, C. J., Conze, D. B., Li, T., Srinivasula, S. M. & Ashwell, J. D. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected]. Nat. Cell Biol. 8, 398–406 (2006).
Salmeron, A. et al. Direct phosphorylation of NF-kappaB1p105 by the IkappaB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J. Biol. Chem. 276, 22215–22222 (2001).
Tojima, Y. et al. NAK is an IkappaB kinase-activating kinase. Nature 404, 778–782 (2000).
Peters, R. T., Liao, S. M. & Maniatis, T. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol. Cell 5, 513–522 (2000).
Helgason, E., Phung, Q. T. & Dueber, E. C. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: the emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 587, 1230–1237 (2013).
Hemmi, H. et al. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199, 1641–1650 (2004).
Perry, A. K., Chow, E. K., Goodnough, J. B., Yeh, W. C. & Cheng, G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J. Exp. Med. 199, 1651–1658 (2004).
Kupka, S. et al. SPATA2-mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep. 16, 2271–2280 (2016).
Komander, D. et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 (2009).
Laplantine, E. et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 28, 2885–2895 (2009).
Ea, C. K., Deng, L., Xia, Z. P., Pineda, G. & Chen, Z. J. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 (2006).
Hadian, K. et al. NF-kappaB essential modulator (NEMO) interaction with linear and Lys-63 ubiquitin chains contributes to NF-kappaB activation. J. Biol. Chem. 286, 26107–26117 (2011).
Chau, T. L. et al. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 33, 171–180 (2008).
Kishore, N. et al. IKK-i and TBK-1 are enzymatically distinct from the homologous enzyme IKK-2: comparative analysis of recombinant human IKK-i, TBK-1, and IKK-2. J. Biol. Chem. 277, 13840–13847 (2002).
Lafont, E. et al. The linear ubiquitin chain assembly complex regulates TRAIL-induced gene activation and cell death. EMBO J. 36, 1147–1166 (2017).
Fitzgerald, K. A. et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 (2003).
Sharma, S. et al. Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151 (2003).
Clement, J. F., Meloche, S. & Servant, M. J. The IKK-related kinases: from innate immunity to oncogenesis. Cell Res. 18, 889–899 (2008).
Clark, K. et al. Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 434, 93–104 (2011).
Schmidt-Supprian, M. et al. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol. Cell 5, 981–992 (2000).
Clark, K., Plater, L., Peggie, M. & Cohen, P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 284, 14136–14146 (2009).
Feng, S. et al. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell. Signal. 19, 2056–2067 (2007).
Lin, Y., Devin, A., Rodriguez, Y. & Liu, Z. G. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 13, 2514–2526 (1999).
O’Donnell, M. A. et al. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat. Cell Biol. 13, 1437–1442 (2011).
Duprez, L. et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 35, 908–918 (2011).
Polykratis, A. et al. Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 193, 1539–1543 (2014).
Pomerantz, J. L. & Baltimore, D. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18, 6694–6704 (1999).
Wagner, S. A., Satpathy, S., Beli, P. & Choudhary, C. SPATA2 links CYLD to the TNF-α receptor signaling complex and modulates the receptor signaling outcomes. EMBO J. 35, 1868–1884 (2016).
Chariot, A. et al. Association of the adaptor TANK with the I kappa B kinase (IKK) regulator NEMO connects IKK complexes with IKK epsilon and TBK1 kinases. J. Biol. Chem. 277, 37029–37036 (2002).
Dynek, J. N. et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 29, 4198–4209 (2010).
Wang, L., Du, F. & Wang, X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell 133, 693–703 (2008).
Peltzer, N., Darding, M. & Walczak, H. Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends Cell Biol. 26, 445–461 (2016).
Annibaldi, A. & Meier, P. Checkpoints in TNF-induced cell death: implications in inflammation and cancer. Trends Mol. Med. 24, 49–65 (2018).
Ting, A. T. & Bertrand, M. J. More to life than NF-kappaB in TNFR1 signaling. Trends Immunol. 37, 535–545 (2016).
Vanden Berghe, T., Kalai, M., van Loo, G., Declercq, W. & Vandenabeele, P. Disruption of HSP90 function reverts tumor necrosis factor-induced necrosis to apoptosis. J. Biol. Chem. 278, 5622–5629 (2003).
Xu, D. et al. TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491 (2018).
Rudolph, D. et al. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 14, 854–862 (2000).
Legarda-Addison, D., Hase, H., O’Donnell, M. A. & Ting, A. T. NEMO/IKKgamma regulates an early NF-kappaB-independent cell-death checkpoint during TNF signaling. Cell Death Differ. 16, 1279–1288 (2009).
Maubach, G., Schmadicke, A. C. & Naumann, M. NEMO links nuclear factor-kappaB to human diseases. Trends Mol. Med. 23, 1138–1155 (2017).
Udalova, I., Monaco, C., Nanchahal, J. & Feldmann, M. Anti-TNF Therapy. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.MCHD-0022-2015 (2016).
Zhao, W. Negative regulation of TBK1-mediated antiviral immunity. FEBS Lett. 587, 542–548 (2013).
Thurston, T. L., Ryzhakov, G., Bloor, S., von Muhlinen, N. & Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221 (2009).
Pilli, M. et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37, 223–234 (2012).
Ahmad, L., Zhang, S. Y., Casanova, J. L. & Sancho-Shimizu, V. Human TBK1: a gatekeeper of neuroinflammation. Trends Mol. Med. 22, 511–527 (2016).
Verhelst, K., Verstrepen, L., Carpentier, I. & Beyaert, R. IkappaB kinase epsilon (IKKepsilon): a therapeutic target in inflammation and cancer. Biochem. Pharmacol. 85, 873–880 (2013).
Vizcaino, J. A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033 (2016).
Acknowledgements
The authors thank all members of the Walczak Group for useful technical advice and fruitful scientific discussions, and A. Betrancourt and J. Kadluczka for excellent technical support. They also thank the following: M. Bertrand for IKKα and IKKβ knockout MEFs; M. Pasparakis for NEMO knockout MEFs; J. Bertin for RIPK1 kinase-dead MEFs; and T. Brooks from the ICH Genetics & Genomic Medicine Programme (London, UK) for RNA-seq services. This work was supported by a Wellcome Trust Investigator Award (096831/Z/11/Z), an ERC Advanced Grant (294880) and a Cancer Research UK programme grant (A17341) awarded to H.W., a Czech Science Foundation grant (17-27355Y) awarded to P.D., and a BBSRC CASE studentship (BB/J013129/1) awarded to M.R. This work also received support from a CRUK–UCL Centre grant (C416/A25145), the CRUK Cancer Immunotherapy Network Accelerator (CITA) Award (C33499/A20265), and the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Author information
Authors and Affiliations
Contributions
H.W. conceived the project. E.L., P.D., E.R., M.R., S.K., A.v.M. and A.L. designed and performed the experiments and analysed the data. S.S., A.B. and K.W. performed the MS experiments and analysed the obtained data. S.S. supervised the MS experiments. H.D. and A.C. performed experiments. D.d.M. generated essential tools for the study. S.H. analysed the RNA-seq data. E.L., E.R., P.D. and H.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.W. is a co‐founder and shareholder of Apogenix AG. All other authors have no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
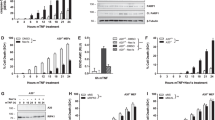
Supplementary Figure 1 LUBAC mediates recruitment of the non-canonical kinases TBK1 and IKKε to and their activation in the TNFR1-SC.
(a) A549 HOIP Control or HOIP KO cells were stimulated with TNF (200 ng/mL) for the indicated times and subjected to lysis, followed by western blot analysis. (b) A549 HOIP Control or HOIP KO cells were stimulated with FLAG-TNF (500 ng/mL) for the indicated times. Lysates and TNFR1-SC were analysed by western blot. (c) HOIP-deficient HeLa cells reconstituted with empty vector, HOIPWT or enzymatically inactive HOIPC885S were stimulated with FLAG-TNF (1 µg/mL) for the indicated time. Lysates and TNFR1-SC were analysed by western blot. (d) HOIP-deficient A549 cells reconstituted with empty vector, HOIPWT or enzymatically inactive HOIPC885S were stimulated with FLAG-TNF (1 µg/mL) for the indicated time. Lysates and TNFR1-SC were analysed by western blot. Unprocessed original scans of blots are shown in Supplementary Figure 7. (a-d) One experiment representative of two independent experiments is shown.
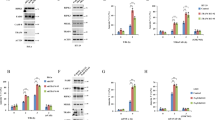
Supplementary Figure 2 Genetic loss or pharmacologic inhibition of TBK1/IKKε exerts only minor effects on TNF-induced gene-activation.
(a,b) MEFs TNF-/- (a) and control or TNF KO; TBK1/IKKε DKO L929 (b) cells were pre-treated with or without the inhibitor MRT as indicated followed by TNF-stimulation (200 ng/mL) for the indicated times and subjected to lysis, followed by western blot analysis. (c) A549 WT cells were pre-treated with either vehicle (DMSO) or the respective inhibitors MRT or TPCA-1 followed by TNF-stimulation (200 ng/mL) for either 0 hr, 1 hr or 4hrs. Three independent experiments for each stimulation time and type of treatment were performed. Cells were then lysed, their total RNA extracted and RNA-Seq analysis performed. The heatmap illustrates the major change of expression across the dataset. The genes selected to be shown were the 100 most highly correlated with PC2 (see Fig 2b). For clarity of comparison the 'rlog' expression data of each row was zeroed at time-point zero hour and then scaled by the standard deviation. The RNA-Seq raw dataset is available in the SRA repository and can be accessed by using the following BioProject accession: PRJNA422567 or SRA accession: SRP126844 (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP126844). (a,b) One experiment representative of two independent experiments is shown. Unprocessed original scans of blots are shown in Supplementary Figure 7.
Supplementary Figure 3 Inhibition or absence of TBK1/IKKε sensitises cells to TNF-induced RIPK1-dependent cell death.
(a, b) MEF TNF-/- (a) and L929 (b) cells were treated with TNF (500 ng/ml) and the indicated compounds. (c) MEF TNF-/- control and MEF TNF-/-; TBK1/ IKKε DKO cells were treated with different doses of TNF for 16 hours. Loss of cell viability was determined using the Cell Titer Glo assay. Mean of three technical replicates from two independent experiments is shown. (d) L929 cells of indicated genotype were stimulated with TNF (50 ng/mL) for 4h. Loss of cell viability was determined using the Cell Titer Glo assay. Mean ± SEM of n=3 independent experiments. (e-j) MEF TNF-/- (e, i, j) and A549 RIPK3-expressing (f-h) cells were stimulated with TNF (500 ng/mL) and pre-treated with or without MRT and BX-795 in combination with zVAD, zVAD/Nec-1s, CHX or zVAD/CHX. (a, b, e-j) Cell death was measured in function of time by SytoxGreen positivity. For all cell death results (a, b, e-j), the RFU mean of 4 technical replicates of one representative experiment out of three independent experiments is represented. Representative images of indicated measurements are depicted with corresponding percentage of dead cells. Cell counting was performed manually using ImageJ. White bar in microscopy images equals 200 µm. Raw data are provided in Supplementary table 1.
Supplementary Figure 4 Inhibition of TBK1/IKKε sensitises primary BMDMs to TNF-induced RIPK1-dependent apoptosis.
(a) Primary BMDMs WT were treated with TNF (100 ng/mL) in the presence or absence of MRT and Nec-1s. (b) Primary BMDMs WT were pre-treated and cultured for two hours with or without TNF blocker ENBREL® (etanercept) (150 µg/mL) and further treated with MRT for 30 minutes. (c-e) Primary BMDMs of the indicated genotype were treated with TNF (100 ng/mL) in the presence or absence of the indicated compounds. (a-e) Cell death was measured in function of time by SytoxGreen positivity. For all cell death results, the mean of SYTOX positive cells per well of 3 technical replicates of one representative experiment out of four independent experiments is represented. Representative images of indicated measurements are depicted with corresponding percentage of dead cells. Cell counting was performed manually using ImageJ. White bar in microscopy images equals 200 µm. Raw data are provided in Supplementary table 1.
Supplementary Figure 5 TBK1 and IKKε phosphorylate RIPK1 both in vitro and in the native TNFR1-SC.
(a) L929 TNF KO control, TBK1 KO, IKKε KO and TBK1/IKKε DKO cells were left untreated or treated with FLAG-TNF (1 µg/mL) and Nec 1s, followed by FLAG-immunoprecipitation. The purified TNFR1-SC was either subjected to USP2 treatment only or concomitant Lambda-phosphatase treatment as indicated. Analysis was performed by western blot. (b) GST-tagged recombinant RIPK1 was incubated with recombinant GST-tagged IKKε in a kinase assay together with Nec-1s. Phosphorylation was revealed by 2D-gel analysis with separation by pI in the first dimension on IPG strips pH 4-7. Second dimension was done via SDS-PAGE. (a,b) One representative experiment out of two independent experiments is shown. Unprocessed original scans of blots are shown in Supplementary Figure 7.
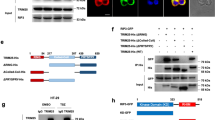
Supplementary Figure 6 NEMO-recruited TANK and NAP1 bring TBK1 and IKKε to the TNFR1-SC.
(a) TANK KO or corresponding control A549 cells and TANK/Optineurin DKO cells were stimulated with FLAG-TNF (500 ng/mL) for the indicated time. The purified TNFR1-SC and lysates were analysed by western blot. (b) A549 cells deficient in TANK, NAP1 or concomitantly deficient in TANK and NAP1 (TANK/NAP1 DKO), or corresponding control cells were stimulated with FLAG-TNF (500 ng/mL) for the indicated time and subjected to immunoprecipitation via Flag. The purified TNFR1-SC and lysates were analysed by western blot. (c) HeLa control and TANK/NAP1 DKO cells were stimulated with FLAG-TNF (500 ng/mL) for the indicated time. The purified TNFR1-SC and lysates were analysed by western blot. (d) WT or NEMO-/- MEFs were treated with FLAG-TNF (500 ng/mL) for the indicated times. The purified TNFR1-SC and lysates were analysed by western blot. (e) MEF WT cells were pre-treated with or without TPCA-1 for 1 hour followed by stimulation with FLAG-TNF (1 µg /mL) for the indicated times and subjected to immunoprecipitation via FLAG. The purified TNFR1-SC and lysates were analysed by western blot. (f) WT and IKKα/β-/-MEF cells pre-treated with or without MRT as indicated were treated with TNF (200 ng/mL) for the indicated times and subjected to lysis, followed by western blot analysis. (g) MEF WT cells were pre-treated with the respective inhibitors and stimulated with TNF (200 ng/mL) for the indicated times. Lysates were analysed by western blot. Unprocessed original scans of blots are shown in Supplementary Figure 7. (h) Schematic summary of the four major findings proposed in this manuscript: (1) In non-stimulated cells TANK constitutively binds to IKKε, while both TANK and NAP1 bind to TBK1. (2) Upon stimulation, the TNFR1-SC is formed, LUBAC is recruited to K63-linkages and creates M1-linkages which in turn recruit NEMO. (3) NEMO, independently of its role in activating IKKα/β, then functions as the docking site for TANK and NAP1, which recruit TBK1 and IKKε to complex I which allows for phosphorylation of RIPK1 on numerous sites. (a-g) One experiment representative of two independent experiments is shown.
Supplementary Figure 7
Uncropped blots
Supplementary information
Supplementary Information
Supplementary Figures 1–7 and Supplementary Table legends.
Supplementary Table 1
Statistics source data.
Supplementary Table 2
Mass spectrometric analysis of LUBAC-mediated recruitment of TNFR1-SC components.
Supplementary Table 3
List of transcripts significantly changed at 1 hour after TNF-stimulation.
Supplementary Table 4
Mass spectrometric analysis of RIPK1 phosphosites.
Supplementary Table 5
Mass spectrometric analysis of the TBK1-interactome.
Supplementary Table 6
List of sequences targeted by the different gRNAs and primers used in this study.
Supplementary Table 7
List of antibodies used in this study.
Rights and permissions
About this article
Cite this article
Lafont, E., Draber, P., Rieser, E. et al. TBK1 and IKKε prevent TNF-induced cell death by RIPK1 phosphorylation. Nat Cell Biol 20, 1389–1399 (2018). https://doi.org/10.1038/s41556-018-0229-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0229-6
This article is cited by
-
Mediators of necroptosis: from cell death to metabolic regulation
EMBO Molecular Medicine (2024)
-
IKKε and TBK1 prevent RIPK1 dependent and independent inflammation
Nature Communications (2024)
-
Immunogenic cell death in cancer: targeting necroptosis to induce antitumour immunity
Nature Reviews Cancer (2024)
-
RIPK3 cleavage is dispensable for necroptosis inhibition but restricts NLRP3 inflammasome activation
Cell Death & Differentiation (2024)
-
LUBAC is required for RIG-I sensing of RNA viruses
Cell Death & Differentiation (2024)