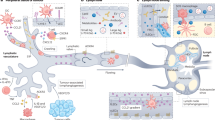
Abstract
Metastatic seeding by disseminated cancer cells principally occurs in perivascular niches. Here, we show that mechanotransduction signalling triggered by the pericyte-like spreading of disseminated cancer cells on host tissue capillaries is critical for metastatic colonization. Disseminated cancer cells employ L1CAM (cell adhesion molecule L1) to spread on capillaries and activate the mechanotransduction effectors YAP (Yes-associated protein) and MRTF (myocardin-related transcription factor). This spreading is robust enough to displace resident pericytes, which also use L1CAM for perivascular spreading. L1CAM activates YAP by engaging β1 integrin and ILK (integrin-linked kinase). L1CAM and YAP signalling enables the outgrowth of metastasis-initiating cells both immediately following their infiltration of target organs and after they exit from a period of latency. Our results identify an important step in the initiation of metastatic colonization, define its molecular constituents and provide an explanation for the widespread association of L1CAM with metastatic relapse in the clinic.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
RNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE82281 and also in Supplementary Table 1. Statistics source data for Figs. 1–8 and Supplementary Figs. 1–7 have been provided in Supplementary Table 1. All the raw images for the western blots can be found in Supplementary Fig. 8. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Change history
12 December 2018
In the version of this Article originally published, the authors inadvertently included the term ‘pericytic mimicry’ in relation to ref. 54. This has now been corrected by inserting an additional reference at position 51 and amending the text in the Discussion relating to ‘pericytic mimicry’, ref. 54 and pericyte-like spreading. The original refs 51–70 have also been renumbered. Furthermore, Fig. 8l has been amended to remove the term ‘pericyte mimicry’ that the authors had included inadvertently during figure preparation. These corrections have been made in the online versions of the Article.
References
Massague, J. & Obenauf, A. C. Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016).
Sosa, M. S., Bragado, P. & Aguirre-Ghiso, J. A. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer 14, 611–622 (2014).
Lambert, A. W., Pattabiraman, D. R. & Weinberg, R. A. Emerging biological principles of metastasis. Cell 168, 670–691 (2017).
Obenauf, A. C. & Massague, J. Surviving at a distance: organ specific metastasis. Trends Cancer 1, 76–91 (2015).
Kienast, Y. et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122 (2010).
Entenberg, D. et al. In vivo subcellular resolution optical imaging in the lung reveals early metastatic proliferation and motility. Intravital 4, e1086613 (2015).
Ritsma, L. et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci. Transl. Med. 4, 158ra145 (2012).
Morrison, S. J. & Scadden, D. T. The bone marrow niche for haematopoietic stem cells. Nature 505, 327–334 (2014).
Rafii, S., Butler, J. M. & Ding, B. S. Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325 (2016).
Malladi, S. et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell 165, 45–60 (2016).
Ghajar, C. M. et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15, 807–817 (2013).
Donnem, T. et al. Vessel co-option in primary human tumors and metastases: an obstacle to effective anti-angiogenic treatment? Cancer Med. 2, 427–436 (2013).
Valiente, M. et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014).
Altevogt, P., Doberstein, K. & Fogel, M. L1CAM in human cancer. Int. J. Cancer 138, 1565–1576 (2016).
Ebeling, O. et al. L1 adhesion molecule on human lymphocytes and monocytes: expression and involvement in binding to αvβ3 integrin. Eur. J. Immunol. 26, 2508–2516 (1996).
Dahme, M. et al. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat. Genet. 17, 346–349 (1997).
Kiefel, H. et al. L1CAM: a major driver for tumor cell invasion and motility. Cell Adhes. Migr. 6, 374–384 (2012).
Armulik, A., Genove, G. & Betsholtz, C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 (2011).
Nguyen, D. X. et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 138, 51–62 (2009).
Bos, P. D. et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009).
Minn, A. J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005).
Shibue, T. & Weinberg, R. A. Integrin β1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc. Natl Acad. Sci. USA 106, 10290–10295 (2009).
Shibue, T., Brooks, M. W. & Weinberg, R. A. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell 24, 481–498 (2013).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Winslow, M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011).
Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P. & Heintz, N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9, 1282–1291 (2014).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Olson, E. N. & Nordheim, A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365 (2010).
Lamar, J. M. et al. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc. Natl Acad. Sci. USA 109, E2441–2450 (2012).
Medjkane, S., Perez-Sanchez, C., Gaggioli, C., Sahai, E. & Treisman, R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11, 257–268 (2009).
Iwamoto, M., Bjorklund, T., Lundberg, C., Kirik, D. & Wandless, T. J. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 17, 981–988 (2010).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Zhao, B., Li, L., Lei, Q. & Guan, K. L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 (2010).
Aragona, M. et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059 (2013).
Elosegui-Artola, A. et al. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 171, 1397–1410 (2017).
Euer, N. I. et al. Identification of L1CAM, Jagged2 and Neuromedin U as ovarian cancer-associated antigens. Oncol. Rep. 13, 375–387 (2005).
McCuskey, R. S., Urbaschek, R. & Urbaschek, B. The microcirculation during endotoxemia. Cardiovasc. Res. 32, 752–763 (1996).
Ziegler, T. et al. Angiopoietin 2 mediates microvascular and hemodynamic alterations in sepsis. J. Clin. Invest. 123, 3436–3445 (2013).
Law, J. W. et al. Decreased anxiety, altered place learning, and increased CA1 basal excitatory synaptic transmission in mice with conditional ablation of the neural cell adhesion molecule L1. J. Neurosci. 23, 10419–10432 (2003).
Zhu, X. et al. Age-dependent fate and lineage restriction of single NG2 cells. Development 138, 745–753 (2011).
Passaniti, A. et al. A simple, quantitative method for assessing angiogenesis and antiangiogenic agents using reconstituted basement membrane, heparin, and fibroblast growth factor. Lab. Invest. 67, 519–528 (1992).
Kogata, N., Tribe, R. M., Fassler, R., Way, M. & Adams, R. H. Integrin-linked kinase controls vascular wall formation by negatively regulating Rho/ROCK-mediated vascular smooth muscle cell contraction. Genes Dev. 23, 2278–2283 (2009).
Abraham, S., Kogata, N., Fassler, R. & Adams, R. H. Integrin β1 subunit controls mural cell adhesion, spreading, and blood vessel wall stability. Circ. Res. 102, 562–570 (2008).
Serrano, I., McDonald, P. C., Lock, F., Muller, W. J. & Dedhar, S. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat. Commun. 4, 2976 (2013).
Thelen, K. et al. The neural cell adhesion molecule L1 potentiates integrin-dependent cell migration to extracellular matrix proteins. J. Neurosci. 22, 4918–4931 (2002).
Byron, A. et al. Anti-integrin monoclonal antibodies. J. Cell Sci. 122, 4009–4011 (2009).
Ferreira, M., Fujiwara, H., Morita, K. & Watt, F. M. An activating β1 integrin mutation increases the conversion of benign to malignant skin tumors. Cancer Res. 69, 1334–1342 (2009).
Pezzella, F. et al. Non-small-cell lung carcinoma tumor growth without morphological evidence of neo-angiogenesis. Am. J. Pathol. 151, 1417–1423 (1997).
Holash, J. et al. Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science 284, 1994–1998 (1999).
Barnhill, R. L. & Lugassy, C. Angiotropic malignant melanoma and extravascular migratory metastasis: description of 36 cases with emphasis on a new mechanism of tumour spread. Pathology 36, 485–490 (2004).
Lugassy, C. et al. Pilot study on ‘‘pericytic mimicry’’ and potential embryonic/stem cell properties of angiotropic melanoma cells interacting with the abluminal vascular surface. Cancer Microenviron. 6, 19–29 (2013).
Breast Cancer Progression Working Party. Evidence for novel non-angiogenic pathway in breast-cancer metastasis. Lancet 355, 1787–1788 (2000).
Sardari Nia, P., Hendriks, J., Friedel, G., Van Schil, P. & Van Marck, E. Distinct angiogenic and non-angiogenic growth patterns of lung metastases from renal cell carcinoma. Histopathology 51, 354–361 (2007).
Stessels, F. et al. Breast adenocarcinoma liver metastases, in contrast to colorectal cancer liver metastases, display a non-angiogenic growth pattern that preserves the stroma and lacks hypoxia. Br. J. Cancer 90, 1429–1436 (2004).
Cheng, L. et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell 153, 139–152 (2013).
Hellstrom, M., Kalen, M., Lindahl, P., Abramsson, A. & Betsholtz, C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047–3055 (1999).
Moroishi, T. et al. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell 167, 1525–1539 e1517 (2016).
Kang, Y. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003).
Vanharanta, S. et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat. Med. 19, 50–56 (2013).
Kasai, M. et al. In vivo effect of anti-asialo GM1 antibody on natural killer activity. Nature 291, 334–335 (1981).
van de Wiel-van Kemenade, E. et al. Adhesion of T and B lymphocytes to extracellular matrix and endothelial cells can be regulated through the beta subunit of VLA. J. Cell Biol. 117, 461–470 (1992).
Lawley, T. J. & Kubota, Y. Induction of morphologic differentiation of endothelial cells in culture. J. Invest. Dermatol. 93, 59S–61S (1989).
Polleux, F. & Ghosh, A. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Science’s STKE: Signal Transduct. Knowl. Environ. 2002, pl9 (2002).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Doyle, J. P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008).
Zhang, X. H. et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 154, 1060–1073 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Hopewell, E. L. et al. Lung tumor NF-κB signaling promotes T cell-mediated immune surveillance. J. Clin. Invest. 123, 2509–2522 (2013).
Mohseni, M. et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat. Cell Biol. 16, 108–117 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Fellmann, C. et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013).
Acknowledgements
We acknowledge members of the MSKCC Molecular Cytology Core and Pathology Core Facilities for their assistance with staining, tissue processing, image acquisition and analysis and Young-Mi Kim for help with YAP activity assays. This work was supported by NIH grants P01-CA094060, P01-CA129243 (J.M.) and P30-CA008748 (Memorial Sloan Ketterin Cancer Center), T32-CA009207 (K.G.), DOD Innovator award W81XWH-12-0074 (J.M.), and the Alan and Sandra Gerry Metastasis Research Initiative (J.M.), Shulamit Katzman Endowed Postdoctoral Research Fellowships (E.E.E. and K.G.), an AACR Basic Cancer Research Fellowship and Conquer Cancer Foundation of ASCO Young Investigator Award (K.G.).
Author information
Authors and Affiliations
Contributions
E.E.E., M.V. and J.M. conceptualized the project. E.E.E., M.V., K.G. and S.M. designed and performed the experiments. Y.Z. performed bioinformatics analyses. S.A. and B.G. assisted with the experiments. F.G.G. contributed to experimental design. M.S. provided the L1camflox mice. E.B., A.B. and M.R. provided clinical samples and interpretation. KG. and M.V. provided critical editing for the manuscript. E.E.E., and J.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Pericytic spreading of metastatic cells.
(a) Representative images of CellTracker+ cells (green) co-cultured with endothelial tubes (bright field) on matrigel (scale bar, 20 μm). (b) Representative images of GFP+ H2030-BrM (white), CD31+ endothelial cells (red) and CSPG4+ pericytes (green) in cultured brain tissue. Right panels are insets. Yellow arrows indicate pericytes 1) displaced, 2) localized across from the cancer cells and 3) stomped on (scale bars, 20 μm). (c) Representative image of a GFP+ MDA231-BrM cell (white) as in b (scale bars, 10 μm). (d-e) Representative immunofluorescence images H2030-BrM cells (GFP, white) and pericytes co-stained with pericyte markers (d) (PDGFRβ, red and CSPG4, green) and oligodendrocyte markers (e) (O4 sulfatide, red and CSPG4, green). Arrowheads: double positive cells (scale bars, 5μm). Figures (a-e) are representative of 2 independent experiments. (f) Representative time lapse confocal images of H2030-BrM control (Ca: white) displacing CSPG4-DsRed+ pericytes (p1, p2 and p3: green) along the endothelium (DiD staining, red) in cultured brain tissue. Right panels, H2030-BrM L1CAM knockdown cells interacting with pericytes (p1-p3, DsRed: green). Arrows: direction of cancer cell movement in the next frame; solid circles: stalling. L1CAM knockdown cells eventually round up. Images are representative of 4 (Control) and 3 (shL1CAM) independent experiments (scale bars, 10 μm). (g) Mean pericyte coverage in cancer cell invested versus adjacent uninvolved vasculature depicted in Fig. 1d (n = 62, 55, 83, 96 vessels per H2030-BrM “+”, “-”, MDA231-BrM “+”, “-” cancer cell groups pooled from 2 independent experiments). P values are calculated using Mann-Whitney test. Error bars, S.E.M. (h) Immunohistochemistry images of metastatic cells (white arrowheads and L1CAM staining) interacting with pericytes and vascular smooth muscle cells (alpha-smooth muscle actin staining, brown) along the vascular wall (red dotted line) in clinically silent micrometastases in brain tissue obtained during autopsy of a treatment-naïve ER+ breast cancer patient (scale bars, 50 μm). (i) Immunofluorescence staining of brain tissue in (h) for pericytes (aSMA staining, green), endothelial cells (CD31 staining, red) and cancer cells (panKeratin staining, white) (scale bar, 5μm). Data in h-i is from one biological replicate. See Supplementary Table 1 for statistics source data.
Supplementary Figure 2 Initiation of metastatic colonization in multiple organs.
(a) Confocal images of GFP+ MDA231-BrM (green) cells spreading along the mouse brain vasculature (lectin staining, red) after infiltrating the brain parenchyma of a mouse via the arterial circulation 7 d after injection (scale bar, 20μm). (b) GFP+ MDA231-LM cells (green) generating vascular coopting protrusions (arrowheads) over alveolar blood capillaries (lectin staining, red) after extravasation from the venous circulation into the lungs of a mouse, 2 d after tail vein injection. Dotted lines outline alveolar space (as) (scale bar, 20μm). (c) L1CAM immunoblot in MDA-MB-468 cells. GAPDH, loading control. (d) Confocal images of MDA-MB-468 cells (CellTracker, green) spreading over the vasculature (Lectin staining, red) in brain slice culture assays (scale bar, 20μm). Experiments in (a-d) are representative of 2 independent experiments. (e) Quantitation of roundness index as an inverse measure of cell spreading in MDA-MB-468 cells expressing control or L1CAM overexpression constructs as in (c-d) (n = 158 and 170 cells pooled from 2 independent experiments). Red lines, median. (f) Representative images of GFP+ MDA231-LM control and L1CAM knockdown cells (green) in lungs harvested from the mice in (a) (scale bar, 100 μm). Data on the right are metastatic lesion size in μm2 (n = 132 and 84 metastatic foci per control and shL1CAM groups pooled from 2 mice each). P-values in (e-f) are calculated by Mann-Whitney test. (g) Representative confocal image of 393N1 mouse metastatic cells (GFP staining, green) colonizing the brain of syngeneic mice and spreading over brain capillaries (CD31 staining, red) (scale bar, 50 μm). Data represent 2 biological replicates. (h) Mean luciferase units of MDA231-BoM, HCT116, 786M1A cells measured by CellTiter-Glo in monolayer cultures in vitro (n = 3 independent experiments in triplicates). Error bars, S.E.M. (i-j) Cell viability (CellTiter-Glo)(i) and cell death (Caspase-Glo)(j) activities in H2030-BrM and MDA231-BrM after they have been cultured for 24 h in suspension (n = 3 independent experiments in triplicate). Data in (i-j) are mean relative luciferase units (RLU) and mean arbitrary units (A.U.). Error bars, S.E.M. See Supplementary Figure 8 for unprocessed blots. See Supplementary Table 1 for statistics source data.
Supplementary Figure 3 L1CAM requirement for metastatic colonization post-extravasation.
(a) Doxycycline inducible L1CAM shRNA lenti-viral vector design. T3G, doxycycline inducible promoter controlling DsRed and L1CAM shRNA expression. PGK, phosphoglycerate kinase promoter for constitutive Venus fluorescent protein expression. (b) Flow cytometry analysis of L1CAM surface expression in the indicated cell lines in the absence (green) or presence (red) of doxycycline. (c) Counter plot representation of data in (c) Red and green scatter plots correspond to red and green histograms in (c). Median intensity: median fluorescence intensity. FSC: forward scatter. (d) Example of the gating strategy used for the flow cytometry experiments in (b-c). (e) Immunohistochemical L1CAM staining showing knockdown efficiency in lungs and brain of metastasis bearing mice. Yellow dotted line separates L1CAM+ brain parenchyma from H2030-BrM lesion with L1CAM knockdown (scale bars, 50 μm). Data in (b-e) are representative of 2 biological replicates. (f) BLI intensity measurements in mice injected with cells carrying doxycycline inducible control vector and treated with or without doxycycline in the diet as in Fig. 3a (n = 8 and 10 mice for control and +dox groups from one experiment with each biological replicate shown). Red lines, median. P values are calculated using Mann-Whitney test. See Supplementary Table 1 for statistics source data.
Supplementary Figure 4 L1CAM requirement for vascular cooption and outgrowth of breast cancer cells exiting from metastatic latency.
(a) Representative images of elongated, rounded, intermediate and clustered (multiple) HCC1954-LCC1 cells found in the brain of mice 9 days after intracardiac injection of control or L1CAM knockdown cells (scale bar, 10μm). Data represent 2 biological replicates per group. Data to the right represent the number of cells belonging to each phenotypic category for control and L1CAM knockdown cells observed in the brains. Lines, mean (n = 72 and 45 cells for control and shL1CAM groups from 2 mice per condition). (b) GFP+ HCC1954-LCC1 cell (green) morphology in the brain microvasculature (lectin staining, red). Top panels: representative images of cells 2 weeks after intracardiac injection with no NK cell depletion. (1) and (2) represent round and spread cells prior to NK cell depletion. Bottom panels: Representative images of cooptive growth of HCC1954-LCC1 cells s 3 weeks after initiation of anti-asialo-GM1 antibody injections to deplete NK cells (scale bars, 10 μm). Data represent 2 (no NK cell depletion) and 5 (three weeks after NK cell depletion) biological replicates. (c) BLI intensity measurements in mice injected with cells carrying doxycycline inducible control vector and treated with or without doxycycline in the diet as in Fig. 4b. n.s., not significant (n = 6 and 5 mice per “–” and “+” groups). Red lines, median. P values are calculated using Mann-Whitney test. Data represent one experiment with each biological replicate shown. (e) Immunoblot images of L1CAM knockdown with doxycycline treatment in HCC1954-LCC1 cells infected with TetON-shL1CAM vector. Data represent 2 independent experiments. (f) Images of Ki67+ (green) HCC1954-LCC1 cells (white) along the vasculature (lectin staining, red) in Fig. 4. Data represent 5 biological replicates per group. See Supplementary Figure 8 for unprocessed blots. See Supplementary Table 1 for statistics source data.
Supplementary Figure 5 L1CAM mediated re-wiring of transcriptional programs.
(a) Heat map of differential gene expression with unsupervised clustering. TRAP RNA sequencing results of H2030-BrM control and L1CAM knockdown cells in BSCs described in Fig. 5a are shown as normalized row z-scores. (b) Top five gene signatures from the TRAP data in (a) ranked based on GSEA normalized enrichment scores (NES). FDR, false discovery rate. Data in (a-b) are from 2 biological replicates. (c) Relative mean expression of YAP target genes ANKRD1, ITGB2, and CTGF in H2030-BrM cells transiently expressing the indicated constructs. YAPWT: YAP Wildtype; YAP5SA: constitutively nuclear YAP mutant; YAPS94A: TEAD binding deficient YAP mutant (n = 4 independent experiments in triplicate). (d) Relative mean expression of MRTF-A target genes in indicated cell lines. MKL1: gene name for MRTF-A. Data are from n = 3 biological replicates. Error bars in (c-d): S.E.M. (e) Fluorescence and bright field images of MDA231-BrM cells in 2D culture 24 hours after addition of small molecule stabilizer with indicated treatments. LatA: Latrunculin A (scale bar, 50μm). Data represent 2 biological replicates. (f) Immunoblots showing RFP protein accumulation in indicated cells with indicated treatments. GFP, loading control. Data represent 2 biological replicates. (g) Immunoblot analysis of phosphorylation of Hippo pathway kinases LATS1 and MST1 in cells plated on collagen for 1 hour. GAPDH loading control. Data represent 3 biological replicates. (h) Mean relative luciferase units in indicated cell lines measured by CellTiter-Glo in monolayer cultures in vitro. Error bars, S.E.M. (n = 3 independent experiments in triplicate). (i) Images of indicated H2030-BrM cells in BSCs (scale bars, 10 μm). Data represent 2 biological replicates. Dot plots: roundness index. Red lines, median (n = 45, 53 and 53 cells for Control, shYAP#1 and shYAP#2 cells pooled from 2 independent experiments). P values are calculated using Mann-Whitney test. (j) YAP immunoblots in indicated cells with or without doxycycline treatment. TetON-shYAP: Doxycycline inducible YAP shRNA construct. Data represent 2 biological replicates. See Supplementary Figure 8 for unprocessed blots. See Supplementary Table 1 for statistics source data.
Supplementary Figure 6 L1CAM regulates YAP nuclear localization.
(a) Mean IHC scoring summary table of serially sectioned and stained patient samples that contain L1CAM+ metastatic cancer cells. Data are from 10 different patients (n = 15, 10, 6, 4, 3, 3, 4, 8, 8, 8 fields of view for patients A-J). Error values are S.D. (b) Single cell image analysis of L1CAM staining intensity with YAP nuclear staining intensity in serially sectioned patient IHC samples. Automated alignment is achieved assigning metastatic cells (L1CAM+, brown circles) to the nearest nucleus in the YAP image (green filled circles). Graphs to the right demonstrate L1CAM staining intensity correlation with YAP staining intensity. P values are calculated by linear regression (n = 159 and 611 cells for upper and lower panels in one field of view shown. Data represent 2 field of views calculated per patient). (c) YAP immunostaining of GFP+ H2030-BrM control and L1CAM knockdown cells on capillaries (Collagen IV staining) in brain tissue cultures depicted in Fig. 6c. Data represent 2 independent experiments. (d) YAP immunostaining of GFP+ MDA231-LM control and L1CAM knockdown cells spreading on vessels in mouse lungs in vivo, 2 days after tail vein injection depicted in Fig. 6d. Cancer cell GFP is cytoplasmic due to its nuclear export signal sequence (scale bars, 10 μm). Data represent 2 biological replicates. (e) Mean relative luciferase units of indicated H2030-BrM and MDA231-BrM cells in vitro, measured by CellTiter-Glo. YAP5SA: constitutively nuclear YAP, YAPS94A: TEAD binding deficient YAP. (n = 3 independent experiments done in three technical replicates). Error bars, S.E.M. See Supplementary Table 1 for statistics source data.
Supplementary Figure 7 L1CAM regulates integrin-ILK signaling.
(a-b) Median Lung and whole body BLI intensities of mice injected with indicated cells and treated as indicated. TetOn-shILK: Doxycycline inducible ILK shRNA construct. (n = 9, 8, 10 and 9 mice per MDA231-LM control, Dox +, MDA231-BoM control and Dox + groups) (c) Representative images of active β1 integrin immunofluorescence (12G10 staining) in Fig. 8d in the indicated H2030-BrM cells plated on indicated substrates.. 12G10 antibody recognizes the β1 integrins specifically in their active confirmation (scale bar, 10 μm). Data represent 2 independent experiments. (d) Immunoblots of Ankyrin B (AnkB) and L1CAM co-immunoprecipitation (IP) in H2030-BrM cells. Input: whole cell lysate Data represent from 2 independent experiments (e) Median integrated active β1 integrin immunofluorescence intensity in indicated cells measured as in Fig. 8d (n = 320 and 268 cells for Control and shANK2 groups pooled from 2 independent experiments). (f) Representative ex vivo BLI images of mouse brains in Fig. 8k (j) (f) (s). In a-b, and e red lines, median and P values are calculated using Mann-Whitney test. Data in a-b are from one experiment with each biological replicate shown See Supplementary Figure 8 for unprocessed blots. See Supplementary Table 1 for statistics source data.
Supplementary Figure 8
Unprocessed western blots.
Supplementary information
Supplementary Information
Supplementary Figures 1–8, and legends for Supplementary Tables 1–2 and Videos 1–4
Rights and permissions
About this article
Cite this article
Er, E.E., Valiente, M., Ganesh, K. et al. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat Cell Biol 20, 966–978 (2018). https://doi.org/10.1038/s41556-018-0138-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0138-8
This article is cited by
-
STAT3–mediated up-regulation of DAB2 via SRC-YAP1 signaling axis promotes Helicobacter pylori-driven gastric tumorigenesis
Biomarker Research (2024)
-
PTEN-restoration abrogates brain colonisation and perivascular niche invasion by melanoma cells
British Journal of Cancer (2024)
-
Evidence and therapeutic implications of biomechanically regulated immunosurveillance in cancer and other diseases
Nature Nanotechnology (2024)
-
Key processes in tumor metastasis and therapeutic strategies with nanocarriers: a review
Molecular Biology Reports (2024)
-
Role of adhesion molecules in cancer and targeted therapy
Science China Life Sciences (2024)