Abstract
Cilia are evolutionarily conserved structures with many sensory and motility-related functions. The ciliary base, composed of the basal body and the transition zone, is critical for cilia assembly and function, but its contribution to cilia diversity remains unknown. Hence, we generated a high-resolution structural and biochemical atlas of the ciliary base of four functionally distinct neuronal and sperm cilia types within an organism, Drosophila melanogaster. We uncovered a common scaffold and diverse structures associated with different localization of 15 evolutionarily conserved components. Furthermore, CEP290 (also known as NPHP6) is involved in the formation of highly diverse transition zone links. In addition, the cartwheel components SAS6 and ANA2 (also known as STIL) have an underappreciated role in basal body elongation, which depends on BLD10 (also known as CEP135). The differential expression of these cartwheel components contributes to diversity in basal body length. Our results offer a plausible explanation to how mutations in conserved ciliary base components lead to tissue-specific diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Bettencourt-Dias, M., Hildebrandt, F., Pellman, D., Woods, G. & Godinho, S. A. Centrosomes and cilia in human disease. Trends Genet. 27, 307–315 (2011).
Jana, S. C., Marteil, G. & Bettencourt-Dias, M. Mapping molecules to structure: unveiling secrets of centriole and cilia assembly with near-atomic resolution. Curr. Opin. Cell Biol. 26, 96–106 (2014).
Ibanez-Tallon, I., Heintz, N. & Omran, H. To beat or not to beat: roles of cilia in development and disease. Hum. Mol. Genet. 12, R27–R35 (2003).
Carvalho-Santos, Z., Azimzadeh, J., Pereira-Leal, J. B. & Bettencourt-Dias, M. Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194, 165–175 (2011).
Newton, F. G. et al. Forkhead transcription factor Fd3F cooperates with Rfx to regulate a gene expression program for mechanosensory cilia specialization. Dev. Cell 22, 1221–1233 (2012).
Yu, X., Ng, C. P., Habacher, H. & Roy, S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 40, 1445–1453 (2008).
Reiter, J. F., Blacque, O. E. & Leroux, M. R. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13, 608–618 (2012).
Kobayashi, T. & Dynlacht, B. D. Regulating the transition from centriole to basal body. J. Cell Biol. 193, 435–444 (2011).
Garcia-Gonzalo, F. R. et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776–784 (2011).
Roberson, E. C. et al. TMEM231, mutated in orofaciodigital and Meckel syndromes, organizes the ciliary transition zone. J. Cell Biol. 209, 129–142 (2015).
Bayless, B. A., Giddings, T. H. Jr, Winey, M. & Pearson, C. G. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell 23, 4820–4832 (2012).
Fliegauf, M., Benzing, T. & Omran, H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893 (2007).
Jana, S. C., Bettencourt-Dias, M., Durand, B. & Megraw, T. L. Drosophila melanogaster as a model for basal body research. Cilia 5, 22 (2016).
Carvalho-Santos, Z. et al. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev. Cell 23, 412–424 (2012).
Gottardo, M., Callaini, G. & Riparbelli, M. G. The cilium-like region of the Drosophila spermatocyte: an emerging flagellum? J. Cell Sci. 126, 5441–5452 (2013).
Gottardo, M. et al. Loss of centrobin enables daughter centrioles to form sensory cilia in Drosophila. Curr. Biol. 25, 2319–2324 (2015).
Chen, J. V. et al. Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila. J. Cell Biol. 211, 435–453 (2015).
Jana, S. C., Mendonca, S., Werner, S. & Bettencourt-Dias, M. Methods to study centrosomes and cilia in Drosophila. Methods Mol. Biol. 1454, 215–236 (2016).
Clare, D. K. et al. Basal foot MTOC organizes pillar MTs required for coordination of beating cilia. Nat. Commun. 5, 4888 (2014).
Martinez-Campos, M., Basto, R., Baker, J., Kernan, M. & Raff, J. W. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165, 673–683 (2004).
Dix, C. I. & Raff, J. W. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759–1764 (2007).
Bouissou, A. et al. γ-Tubulin ring complexes regulate microtubule plus end dynamics. J. Cell Biol. 187, 327–334 (2009).
Blachon, S. et al. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics 182, 133–144 (2009).
Muhlhans, J., Brandstatter, J. H. & Giessl, A. The centrosomal protein pericentrin identified at the basal body complex of the connecting cilium in mouse photoreceptors. PLoS ONE 6, e26496 (2011).
Muresan, V., Joshi, H. C. & Besharse, J. C. γ-Tubulin in differentiated cell types: localization in the vicinity of basal bodies in retinal photoreceptors and ciliated epithelia. J. Cell Sci. 104, 1229–1237 (1993).
Liang, A. et al. γ-Tubulin is permanently associated with basal bodies in ciliates. Eur. J. Cell Biol. 70, 331–338 (1996).
Arikawa, K. & Williams, D. S. Acetylated α-tubulin in the connecting cilium of developing rat photoreceptors. Invest. Ophthalmol. Vis. Sci. 34, 2145–2149 (1993).
Galletta, B. J. et al. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 7, 12476 (2016).
Dzhindzhev, N. S. et al. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24, 2526–2532 (2014).
Guichard, P., Chretien, D., Marco, S. & Tassin, A. M. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 29, 1565–1572 (2010).
Gonzalez, C., Tavosanis, G. & Mollinari, C. Centrosomes and microtubule organisation during Drosophila development. J. Cell Sci. 111, 2697–2706 (1998).
Craige, B. et al. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190, 927–940 (2010).
Li, C. et al. MKS5 and CEP290 dependent assembly pathway of the ciliary transition zone. PLoS Biol. 14, e1002416 (2016).
Rachel, R. A. et al. CEP290 alleles in mice disrupt tissue-specific cilia biogenesis and recapitulate features of syndromic ciliopathies. Hum. Mol. Genet. 24, 3775–3791 (2015).
Baker, J. D., Adhikarakunnathu, S. & Kernan, M. J. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development 131, 3411–3422 (2004).
Enjolras, C. et al. Drosophila Chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J. Cell Biol. 197, 313–325 (2012).
Basiri, M. L. et al. A migrating ciliary gate compartmentalizes the site of axoneme assembly in Drosophila spermatids. Curr. Biol. 24, 2622–2631 (2014).
Barker, A. R., Renzaglia, K. S., Fry, K. & Dawe, H. R. Bioinformatic analysis of ciliary transition zone proteins reveals insights into the evolution of ciliopathy networks. BMC Genomics 15, 531 (2014).
Schouteden, C., Serwas, D., Palfy, M. & Dammermann, A. The ciliary transition zone functions in cell adhesion but is dispensable for axoneme assembly in C. elegans. J. Cell Biol. 210, 35–44 (2015).
Yang, T. T. et al. Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci. Rep. 5, 14096 (2015).
Pratt, M. B. et al. Drosophila sensory cilia lacking MKS proteins exhibit striking defects in development but only subtle defects in adults. J. Cell Sci. 129, 3732–3743 (2016).
Vieillard, J. et al. Transition zone assembly and its contribution to axoneme formation in Drosophila male germ cells. J. Cell Biol. 214, 875–889 (2016).
Drivas, T. G., Holzbaur, E. L. & Bennett, J. Disruption of CEP290 microtubule/membrane-binding domains causes retinal degeneration. J. Clin. Invest. 123, 4525–4539 (2013).
Singh, H. et al. Visualization and quantification of cardiac mitochondrial protein clusters with STED microscopy. Mitochondrion 12, 230–236 (2012).
Rodrigues-Martins, A. et al. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17, 1465–1472 (2007).
Lin, Y. C. et al. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 32, 1141–1154 (2013).
Guichard, P. et al. Cell-free reconstitution reveals centriole cartwheel assembly mechanisms. Nat. Commun. 8, 14813 (2017).
Basto, R. et al. Flies without centrioles. Cell 125, 1375–1386 (2006).
Laurencon, A. et al. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 8, R195 (2007).
Cachero, S. et al. The gene regulatory cascade linking proneural specification with differentiation in Drosophila sensory neurons. PLoS Biol. 9, e1000568 (2011).
Kong, D. et al. Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J. Cell Biol. 206, 855–865 (2014).
Riparbelli, M. G., Colozza, G. & Callaini, G. Procentriole elongation and recruitment of pericentriolar material are downregulated in cyst cells as they enter quiescence. J. Cell Sci. 122, 3613–3618 (2009).
Doroquez, D. B., Berciu, C., Anderson, J. R., Sengupta, P. & Nicastro, D. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. eLife 3, e01948 (2014).
Soley, J. T. A comparative overview of the sperm centriolar complex in mammals and birds: variations on a theme. Anim. Reprod. Sci. 169, 14–23 (2016).
Hilbert, M. et al. SAS-6 engineering reveals interdependence between cartwheel and microtubules in determining centriole architecture. Nat. Cell Biol. 18, 393–403 (2016).
Saurya, S. et al. Drosophila Ana1 is required for centrosome assembly and centriole elongation. J. Cell Sci. 129, 2514–2525 (2016).
Franz, A., Roque, H., Saurya, S., Dobbelaere, J. & Raff, J. W. CP110 exhibits novel regulatory activities during centriole assembly in Drosophila. J. Cell Biol. 203, 785–799 (2013).
Martin, C. A. et al. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat. Genet. 46, 1283–1292 (2014).
Szymanska, K. et al. Founder mutations and genotype–phenotype correlations in Meckel–Gruber syndrome and associated ciliopathies. Cilia 1, 18 (2012).
Abe, S., Asakura, S. & Ukeshima, A. Formation of flagella during interphase in secondary spermatocytes from Xenopus laevis in vitro. J. Exp. Zool. 246, 65–70 (1988).
Burton, P. R. Ultrastructural studies of microtubules and microtubule organizing centers of the vertebrate olfactory neuron. Microsc. Res Tech. 23, 142–156 (1992).
Fox, H. & Hamilton, L. Ultrastructure of diploid and haploid cells of Xenopus laevis larvae. J. Embryol. Exp. Morphol. 26, 81–98 (1971).
Keil, T. A. Sensory cilia in arthropods. Arthropod Struct. Dev. 41, 515–534 (2012).
Jana, S. C., Girotra, M. & Ray, K. Heterotrimeric kinesin-II is necessary and sufficient to promote different stepwise assembly of morphologically distinct bipartite cilia in Drosophila antenna. Mol. Biol. Cell 22, 769–781 (2011).
Arama, E., Agapite, J. & Steller, H. Caspase activity and a specific cytochrome c are required for sperm differentiation in Drosophila. Dev. Cell 4, 687–697 (2003).
Albert, J. T. & Gopfert, M. C. Hearing in Drosophila. Curr. Opin. Neurobiol. 34, 79–85 (2015).
Mastronarde, D. N. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343–352 (1997).
Acknowledgements
We thank T. Avidor-Reiss, B. Durand, T. Megraw and J. Raff for reagents. We thank B. Durand, P. Bastin, A. Dammermann, L. Saúde, J. Shah, G. Marteil, M. Lince Faria, S. Zitouni and MBD Lab members for reviewing the manuscript and providing helpful discussions on the manuscript. We thank the Gurdon Institute Imaging Facility (Cambridge, UK), the Bacterial Cell Biology Lab (M. Pinho, ITQB, Portugal), IGC Advance Imaging (and its Head, G G. Martins), Histopathology and Electron Microscopy units (A. L. Sousa, S. Bonucci and E. Tranfield) for helping with sample preparation and image acquisition, and the IGC fly facility for fly husbandry. S.C.J. and S.W. are supported by the FCT (Fundação Portuguesa para a Ciência e Tecnologia) Fellowships SFRH/BPD/87479/2012 and SFRH/BD/52176/2013, respectively. The laboratory of H.M. is supported by the ERC (ERC-681443-CODECHECK) and FLAD Life Science 2020. M.B.-D. and her laboratory are supported by the Fundação Calouste Gulbenkian/Instituto Gulbenkian de Ciência, an EMBO installation grant and ERC grants (ERC-261344-CentrioleStructNumb and ERC-683258-CentrioleBirthDeath).
Author contributions
S.C.J., S.M. and P.M. performed most of the experiments with assistance from J.R., S.W. and A.P. S.W. cloned the CEP290 from the fly, while S.C.J. characterized the transgenic flies. S.C.J. performed the walking behaviour experiments and STED experiments with help from J.R., and A.P. and H.M., respectively. S.C.J. and M.B.-D. designed all of the experiments with input from S.M., P.M., S.W., A.P. and H.M. S.C.J. and M.B.-D. wrote the manuscript and all authors commented on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Quantification of the localisation patterns of ciliary base components in different ciliated cells in adult Drosophila.
a, b) Scheme of different Drosophila ciliated cells that were studied in this work. a) Large basiconic olfactory and auditory neurons located in the marked regions of the third and second segment of the adult antenna, respectively. Primary cilia-like structure in spermatocytes and long flagella in early elongating spermatid were also studied. Schemes show longitudinal views and representative electron micrograph cross-sections of the marked regions. b) The scheme represents basal body (BB) elongation and diverse cilia assembly during Drosophila spermatogenesis. c, d) Example wide-field fluorescence and 3D-SIM micrographs of olfactory and spermatocyte cilia. Scheme of a set of three olfactory neurons innervating sensillum basiconica (c) and spermatocyte (d) showing BBs and cilia. Representative wide-field pictures (ii) and SIM pictures (iii) showing acetylated α-tubulin and PACT, a BB marker, in a set of three olfactory neurons. e) Representative wide-field pictures of olfactory (i) and auditory (ii) cilia showing rootletin (red), SAS4 (green) and DPLP (blue), a PCM marker. Arrow heads marks the rootlet region at the ciliary base. The scale bars in a, c, d and e are 100 nm, 10 µm, 10 µm and 1 µm, respectively. f-m) Quantification of the localisation patterns of BB, PCM and TZ components in neurons and sperm cells obtained using 3D-SIM. i) Scheme shows the olfactory, auditory, spermatocyte and spermatid ciliary bases with PCM and BBs (f-i) and TZs (j-m). ii-iii) Schemes show the method for quantification of proteins and different parameters. iv) Length (with mean ± S.D.), diameter (mean ± S.D.) and other variables (mean ± S.D.) of the defined zones are mentioned in the table. All values mentioned in the table are in nanometer (nm). n = 16 samples. The schemes (in left) representing the localisation patterns of the proteins are drawn based on the quantification shown in the right. The experiments presented in a, c, d, and f were repeated independently thrice, while the experiments in e were repeated independently twice. Notably, we used both TEM of ciliary bases, and 3D-SIM of a common BB marker, PACT (the C-terminal domain of pericentrin-like protein (PLP)) to estimate BB size independently, and obtained concordant results (see Fig. 1a–d, Supplementary Fig. 1F-I,2).
Supplementary Figure 2 Different ciliary bases show both similar and variable elements.
a) Left: Longitudinal tomogram stills of the ciliary base in olfactory neurons showing a proximal BB (pBB-white arrowhead), a distal BB (dBB- red arrowhead) and the transition zone (TZ). Right: Model based on the tomogram data. Model of the BB and ciliary microtubules (light green), cytoplasmic MTs (orange) that nucleate from the BBs, MTs that nucleate from the proximal BB and extend into the cilia (brown), non-MT electron densities around BBs (dark blue), the electron densities of rootlet (cyan), vesicles at the ciliary base (magenta), connections between dBB-cell membrane (golden) and the cell/ciliary membranes (black) (see also Fig. 1 and Supplementary Video V1). Based on features at the ciliary base, olfactory neurons can be divided into two types: Type-1) where singlet MTs are absent in the lumen of both BB and TZ (example 1) and Type-2) where one or more singlet MTs (orange) are present in the lumen of BBs and TZ (example 2). ~70% of olfactory neurons are of Type-1, while ~30% of them are of Type-2 (quantification not shown). b) Longitudinal tomogram stills of the ciliary base in auditory neurons showing pBB (white arrowhead), dBB (red arrowhead) and TZ. Right: Model based on the tomogram data. For auditory neurons, we modelled all objects described in a, electron density around the MTs in TZ (dark blue), and the rootlet striations (magenta). Notably, in the example 2 of olfactory cilia (a) we observed singlet microtubules (white arrows) that are bent in olfactory neurons, and the connections between the dBB and the cell membrane are less obvious in single sections of both types of neurons, justifying the importance of collecting and analysing tomograms to model these ciliary bases. c) Electron micrographs of different types of bases showing cytoplasmic MTs around BB and rootlets. The insets present regions marked with dotted squares. d, e) Representative electron micrographs of longitudinal (i) and sets of serial cross sections of marked regions (ii) of BBs in olfactory (d) and auditory neurons (e). For the cross-section series analysis, 70 nm serial sections were collected. Note that the number of singlets and the relative position of singlets and doublets in the pBB varies between individual auditory neurons. All electron micrographs in c-e represent features observed in 3 samples (the experiments presented in this figure were repeated independently twice with similar results). Scale bars on the longitudinal (a, b, c, di and fi) and cross (dii and fii) section micrographs are 500 nm and 100 nm, respectively.
Supplementary Figure 3 Both length and non-MT based electron-dense structures of the TZ vary between different cilia types.
Representative electron micrographs show longitudinal sections (i) and sets of serial cross sections (ii) of the marked regions in the TZs of olfactory neurons (a), auditory neurons (b), spermatocytes (c) and early elongating spermatids (d). For cross-section series analysis, 70 nm serial sections were collected. Arrowheads mark the transition fibres (similar to distal appendage) that connect the distal tip of the BB to the ciliary membrane. The white arrow marks the single MT in the lumen of the spermatid BB in d. The cross section images in a, c and d are from serial sections of a TZ of olfactory, spermatocyte and spermatid, respectively. The cross section images in b were compiled from three different sets of serial sections. Notably, here the area that is distal to the BB and shows electron density on and around the MTs in longitudinal sections of ciliary bases is considered TZ. The region distal to the TZ is considered ciliary shaft (axoneme). All electron micrographs shown here represent features that were observed in 3 samples (the experiments presented in this figure were repeated independently twice with similar results). Scale bars on the longitudinal (a-di) and cross (a-dii) section micrographs are 500 nm and 100 nm, respectively (see also Fig. 3 and Supplementary Videos V7,8).
Supplementary Figure 4 CEP290 localises to the TZ in 9-fold symmetric manner and is required to form cilia in all Drosophila ciliated cells.
a-c) Localisation analysis of GFP tagged CEP290 proteins at different TZs. a) Representative SIM images of the ciliary bases of olfactory neurons marked using acetylated α-tubulin (red) and ectopically expressing CEP290::GFP (Gal4cha19b/UAS-CEP290::GFP) or GFP::CEP290 (Gal4cha19b/UAS-GFP::CEP290). SIM analysis shows CEP290::GFP localises towards the lumen and on the MTs and GFP::CEP290 localises towards the ciliary membrane in the olfactory TZ. b) Schemes show the method of measuring the distance (d) between the MT or the hook and ciliary membrane at the different TZs. c) (i) Schemes show methods of measuring the inter-distance between the outer (do) and inner (di) tips of adjacent MT-membrane linkers. Representative STED micrographs of GFP::CEP290 in the cross-section of the TZs of olfactory (i: do = 99 ± 23 nm; n = 194), auditory (ii: do = 100 ± 24 nm; n = 59) and spermatocyte (iii: do = 90 ± 23 nm; n = 112) cilia. While the white arrowheads mark the resolved GFP foci, empty arrowheads (with dotted border) indicate the postulated missing foci´s position. d-h) CEP290 is required for all cilia assembly in the fly. d) i) The scheme shows the odour repulsion test using the T-tube to measure the ability of adult flies to detect a repulsive odour (Benzaldehyde). ii) Quantification of the percentage of flies that are in the compartment with repulsive odor. A null mutant of Orco, a co-receptor essential for olfaction, was used as a positive control (control1, n = 60 and Orcomutant, n = 60: **p < 0.0001, two-tail Mann-Whitney Test; control2, n = 80 and CEP290RNAi1, n = 90: **p < 0.0001, two-tail Mann-Whitney Test). e) i) Scheme depicts the bang assay and the vertical tube used to test the gravitaxis ability of adult flies. ii) Quantification of the time taken by ≥80% of the flies to successfully climb the half height mark of the tube (18 cm long) (control1, n = 70, control2, n = 60 and CEP290RNAi1, n = 60: **p < 0.0001, two-tail Mann-Whitney Test). iii) Representative kymographs of ten flies with respective genotypes followed for the first 5 seconds after the bang. f) i) Representative pictures of olfactory cilia (marked using Acetylated α-Tubulin) in flies with different genotypes. ii) Representative images of different types of olfactory shafts. iii) Quantification of ciliary defects in flies with different genotypes (control1, n = 36, Cep290mutant, n = 27, and CEP290RNAi1, n = 25). g) i) Representative electron micrographs of cross sections of scolopale in second antennal segments of flies with different genotypes. ii) Quantification of percentage of scolopale with two or more cilia in flies with different genotypes (control1, n = 48, and CEP290RNAi1, n = 34). h) i) Quantification of number of progeny produced per male with different genotypes (control1, n = 10, control3, n = 11, and CEP290RNAi2, n = 11: **p < 0.0001, two-tail Mann-Whitney Test). ii) Representative pictures of the BBs in spermatids marked using BLD10, a centriolar protein, in flies with different genotypes. DNA, BB and sperm flagella are marked by DAPI (blue), BLD10 (green) and acetylated tubulin (red), respectively. iii) Quantification of the length of BBs marked using BLD10 as shown in (ii) (control3, n = 153, and CEP290RNAi2, n = 90: **p < 0.0001, two-tail Mann-Whitney Test). iv) Representative cross-section micrographs show the axoneme bundle of the elongating flagella in different flies. Notably, while 9 + 2 arrangement of the MTs was normal in control3 flies, the MT arrangement was defective in CEP290RNAi2 flies (see insets). All experiments were repeated independently with similar results: a (thrice), c (twice), d (thrice), e (thrice), f (twice), g (twice), h (twice). Scale bars in a, c, eiii, f, g, hii and Hiv represent 1 µm, 100 nm, 1 cm, 10 µm, 500 nm, 10 µm and 500 nm, respectively. In each Tukey-box plot, centre line indicates median and error bars indicate full range of variation (from minimum-to-maximum) and dots are outliers. For different variables of each Tukey-box plot and fly genotypes see Supplementary Table 4 and Supplementary Tables 1,3, respectively.
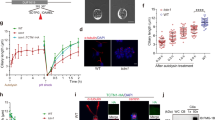
Supplementary Figure 5 Controls for the specificity of RNAi tools used in this manuscript.
Tukey-Box plot of the number of progeny produced per male (i), length of BBs (ii) and total GFP intensity of candidate proteins at the spermatocyte ciliary base: CEP290 (a), SAS6 (b), ANA2 (c) and BLD10 (d). Here, for each candidate (X) gene S1, S2, S3 and S4 represent the flies with ectopic-expression of UAS-mCD8GFP (Control 1X), RNAi (Knock downX), UAS-X-GFP in RNAi background (RescueX) and UAS-X-GFP in wild type background (Control 2X(cOE)). cOE indicates conditional over-expression of the given candidate in the testes. While for a-di the total number of males used for each genotype is n ≥ 10, for a-dii and b-diii, the number of BBs quantified for each genotype is n ≥ 63 and n ≥ 60 (30 pairs of BBs), respectively. For Aiii, the number of BB quantified for each genotype is n ≥ 42. NA indicates not applicable. Note that we rescued the knock down phenotypes of all candidate molecules (SAS6, ANA2, CEP290, and BLD10) both for BB length (ii) and male fertility (i). We further quantified the protein depletion (iii) in RNAi experiments in sperm cells for all candidates. Altogether, this analysis shows the specificity of the tools we used. All experiments presented in a-d were repeated independently twice with similar results. In each Tukey-box plot, centre line indicates median and error bars indicate full range of variation (from minimum-to-maximum) and dots are outliers. For exact sample size, different variables of each Tukey-box plot, statistical tests used, exact p values and fly genotypes see Supplementary Table 4 and Supplementary Tables 1,3, respectively.
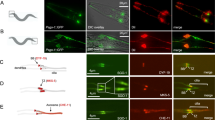
Supplementary Figure 6 SAS6 is differentially required in neurons and sperm cells.
a, b) SAS6 is essential for centriole assembly in neurons, but is not required for neuronal cilia function. a) Representative electron micrographs show longitudinal and cross sections through the centrioles in olfactory neurons (before ciliogenesis: at 24 h APF) in wild type flies. Note that those centrioles are close to the cell membrane and have the cartwheel. b) i) The experimental setting used to reduce/remove SAS6 during centriole and cilia biogenesis in neurons. ii) Representative images show olfactory and auditory neurons in flies with different genotypes. Cilia in olfactory and auditory neurons were studied using anti-acetylated tubulin (green) and anti-glutamylated tubulin (green) antibody, respectively. PLP (red, centrosomes) and DAPI (blue, DNA). Arrowheads mark BBs and arrows mark cilia. c) Both SAS6 and ANA2 are required for sperm BB elongation, being important for male fertility. i) Schematic representation of the experimental setting used to reduce/remove SAS6 before and after centriole biogenesis in sperm cells. ii) Tukey-Box plot of the number of progeny produced per male with different genotypes (control1, n = 10, control5, n = 10, and SAS6RNAi2, n = 11: **p < 0.001, two-tail Mann-Whitney Test; control3, n = 11, SAS6RNAi3, n = 10 **p < 0.01, ANA2RNAi1, n = 10 *p < 0.05, and BLD10RNAi1, n = 10: **p < 0.01, Kruskal-Wallis Test). iii) Tukey-Box plot of the number of BBs per cell in mature spermatocytes (control1, n = 32, and SAS6mutant, n = 42: **p < 0.0001, two-tail Mann-Whitney Test; control3, n = 62, SAS6RNAi3, n = 56, ANA2RNAi1, n = 40, and BLD10RNAi1, n = 63). iv) Representative images of mature spermatocyte BBs of flies with different genotypes (control3, SAS6RNAi3 and ANA2RNAi1). RFP::PACT (red) marks BBs and Anti-SAS6 antibody (green) stains the proximal part of the centriole. Insets show SAS6 (green) close to the arrowhead (in grey scale). v) Tukey-Box plot of the total amount of SAS6 at the mature spermatocyte BBs of the different genotypes. For v (control3, n = 60, SAS6RNAi3, n = 42 **p < 0.0001, and ANA2RNAi1, n = 40, **p < 0.0001: Kruskal-Wallis Test; n represent the number of BB pairs). All experiments in a, cii, ciii were repeated independently twice, while in b, civ-v were repeated independently thrice with similar results. Scale bars in a, bii,iii and civ represent 100 nm, 5 µm and 1 µm, respectively. In each Tukey-box plot, centre line indicates median and error bars indicate full range of variation (from minimum-to-maximum) and dots are outliers. For different variables of each Tukey-box plot and fly genotypes see Supplementary Table 4 and Supplementary Tables 1,3, respectively.
Supplementary Figure 7 SAS6 and ANA2 cooperate to elicit ectopic neuronal BB elongation, leading to defects in sensory behaviour.
a) Representative electron micrographs of longitudinal sections through the ciliary base of olfactory neurons in flies with different genotypes. Empty arrow head marks the proximal region of the dBB without cartwheel. b) Representative electron micrographs of the dBB cross sections of olfactory neurons in flies with either no or simultaneous ectopic expression of SAS6 and ANA2. c) Quantification of the time taken by ≥80% of the flies to successfully climb the half way mark of the 18 cm long tube (SAS6::GFP/GFP::ANA2: −/−, n = 70, +/−, n = 80: (ns-not significant)p > 0.05, −/+, n = 70: (ns)p > 0.05, and +/+, n = 60: **p < 0.0001, Kruskal-Wallis Test). d) The method of measuring the dBB length (LBB). e) Representative SIM images present the longitudinal view of the BBs in flies with different genotypes. PACT (red) marks the neuronal BBs. Insets show PACT (in grey scale) close to the arrowhead. Quantification of BB length (ii), olfactory reception (iii) and gravitaxis (iv) behaviour in flies with either no or ectopic expression of both SAS6 and ANA2 using a driver that only expresses in pre-neuronal cells (Gal4neur). In ii (SAS6::GFP/GFP::ANA2: −/−, n = 47, and +/+, n = 45: **p < 0.0001, two-tail Mann-Whitney Test), iii (SAS6::GFP/GFP::ANA2: −/−, n = 70, and +/+, n = 60: **p < 0.0001, two-tail Mann-Whitney Test) and iv (SAS6::GFP/GFP::ANA2: −/−, n = 60, and +/+, n = 60: **p < 0.0001, two-tail Mann-Whitney Test). Note that upon ectopic expression of both SAS6 and ANA2 in pre-neuronal cells, we observed one cilium per cell, in most of the olfactory neurons (98%: shown in figure), as observed in controls. In the remainder olfactory neurons (not shown), we observed that cells were forming two cilia, each with one BB. (v) Representative electron micrographs show longitudinal sections through the ciliary base and the distal BB cross sections of olfactory neurons in flies with different genotypes. The empty arrow head marks the proximal region of the dBB without cartwheel. Electron micrographs of the dBB cross sections present the features (including 9-fold symmetric doublet MTs) of n ≥ 7 neurons for each genotype. vi) Tukey-Box plot of dBB length in olfactory neurons (SAS6::GFP/GFP::ANA2: −/−, n = 7, and +/+, n = 8: **p < 0.0001, two-tail Mann-Whitney Test). EM (a, b, dv-vi), SIM (ei-ii), and behaviour (c, eiii-iv) experiments were repeated independently twice, thrice and thrice with similar results, respectively. Scale bars in a, b, ei, and ev represent 0.5, 0.2, 1 and 0.1 µm, respectively. In each Tukey-box plot, centre line indicates median and error bars indicate full range of variation (from minimum-to-maximum). For different variables of each Tukey-box plot and fly genotypes see Supplementary Table 4 and Supplementary Tables 1,3, respectively.
Supplementary Figure 8 SAS6 and ANA2 recruit BLD10/CEP135.
a) Schematic representation of the experimental setting used to ectopically express SAS6 and ANA2 after BB docking in the neurons. b) Representative images of neuronal BBs and cell bodies of flies with different genotypes. We analysed RFP::PACT (red) that marks BBs and some centrosomal components, such as Anti-BLD10 and ANA1 antibodies (blue). Insets marked 1 show the region around BBs, while insets marked 2 show cell bodies with accumulation of GFP (either SAS6 or ANA2 or both of them). These experiments were repeated independently thrice displaying similar results. Scale bars represent 1 µm.
Supplementary information
Supplementary Information
Supplementary Figures 1–8, Supplementary Table and Supplementary Video legends, and Supplementary References
Supplementary Table 1
Detailed information about the flies used in this study
Supplementary Table 2
List of antibodies and their dilutions used in this study
Supplementary Table 3
Description of the genotypes of the flies that were used in this study
Supplementary Table 4
Source data
Supplementary Video 1
Structures of the ciliary base in olfactory neurons
Supplementary Video 2
Structures of the auditory ciliary base
Supplementary Video 3
The proximal basal body of olfactory neurons is composed of radially symmetric nine MT doublets
Supplementary Video 4
Nine radially symmetric MT doublets are found in the olfactory distal basal body
Supplementary Video 5
The proximal basal body of auditory neurons is composed of a mixture of nine MT singlets and doublets
Supplementary Video 6
Nano-structures in the distal basal body of auditory neurons are nine-fold symmetric
Supplementary Video 7
Nine-fold symmetric nano-structures are found in the olfactory transition zone
Supplementary Video 8
The transition zone of auditory neurons is composed of nine radially symmetric MT doublets and nano-structures
Rights and permissions
About this article
Cite this article
Jana, S.C., Mendonça, S., Machado, P. et al. Differential regulation of transition zone and centriole proteins contributes to ciliary base diversity. Nat Cell Biol 20, 928–941 (2018). https://doi.org/10.1038/s41556-018-0132-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0132-1
This article is cited by
-
Transport and barrier mechanisms that regulate ciliary compartmentalization and ciliopathies
Nature Reviews Nephrology (2024)
-
GPRC5C regulates the composition of cilia in the olfactory system
BMC Biology (2023)
-
Primary cilia as dynamic and diverse signalling hubs in development and disease
Nature Reviews Genetics (2023)
-
Three Novel Variants of CEP290 and CC2D2DA and a Link Between ZNF77 and SHH Signaling Pathway Are Found in Two Meckel-Gruber Syndrome Fetuses
Reproductive Sciences (2022)
-
Structure and dynamics of photoreceptor sensory cilia
Pflügers Archiv - European Journal of Physiology (2021)