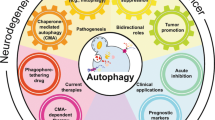
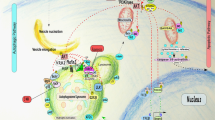
Abstract
The field of autophagy research has developed rapidly since the first description of the process in the 1960s and the identification of autophagy genes in the 1990s. Autophagy is now increasingly studied at the level of organismal pathophysiology and is being connected to the medical sciences. This Historical Perspective describes a brief history of autophagy and discusses unanswered cell biological questions in the field.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ktistakis, N. T. In praise of M. Anselmier who first used the term “autophagie” in 1859. Autophagy 13, 2015–2017 (2017).
Klionsky, D. J. Autophagy revisited: a conversation with Christian de Duve. Autophagy 4, 740–743 (2008).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Stolz, A., Ernst, A. & Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16, 495–501 (2014).
Gatica, D., Lahiri, V. & Klionsky, D. J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242 (2018).
Mizushima, N., Levine, B., Cuervo, A. M. & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008).
Tekirdag, K. A. & Cuervo, A. M. Chaperone-mediated autophagy and endosomal microautophagy: joint by a chaperone. J. Biol. Chem. http://doi.org/cmzv (2017).
Mortimore, G. E. & Pösö, A. R. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 7, 539–564 (1987).
Meijer, A. J. & Codogno, P. Regulation and role of autophagy in mammalian cells. Int. J. Biochem. Cell Biol. 36, 2445–2462 (2004).
Kovács, A. L., Pálfia, Z., Réz, G., Vellai, T. & Kávacs, J. Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy 3, 655–662 (2007).
Biazik, J., Vihinen, H., Anwar, T., Jokitalo, E. & Eskelinen, E. L. The versatile electron microscope: an ultrastructural overview of autophagy. Methods 75, 44–53 (2015).
Tsukada, M. & Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 (1993).
Thumm, M. et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280 (1994).
Harding, T. M., Morano, K. A., Scott, S. V. & Klionsky, D. J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602 (1995).
Yuan, W., Tuttle, D. L., Shi, Y. J., Ralph, G. S. & Dunn, W. A. Jr. Glucose-induced microautophagy in Pichia pastoris requires the α-subunit of phosphofructokinase. J. Cell Sci. 110, 1935–1945 (1997).
Sakai, Y., Koller, A., Rangell, L. K., Keller, G. A. & Subramani, S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J. Cell Biol. 141, 625–636 (1998).
Mukaiyama, H. et al. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes. Cell 7, 75–90 (2002).
Titorenko, V. I., Keizer, I., Harder, W. & Veenhuis, M. Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J. Bacteriol. 177, 357–363 (1995).
Klionsky, D. J. et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545 (2003).
Parzych, K. R., Ariosa, A., Mari, M. & Klionsky, D. J. A newly characterized vacuolar serine carboxypeptidase, Atg42/Ybr139w, is required for normal vacuole function and the terminal steps of autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell http://doi.org/cmpc(2018).
Nakatogawa, H., Suzuki, K., Kamada, Y. & Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 (2009).
Tian, Y. et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055 (2010).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011).
Muller, O. et al. Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 151, 519–528 (2000).
Mukaiyama, H. et al. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 15, 58–70 (2004).
Krick, R. et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 19, 4492–4505 (2008).
Wang, C. W., Miao, Y. H. & Chang, Y. S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206, 357–366 (2014).
van Zutphen, T. et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 25, 290–301 (2014).
Tsuji, T. et al. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. eLife 6, e25960 (2017).
Sahu, R. et al. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20, 131–139 (2011).
Liu, X. M. et al. ESCRTs cooperate with a selective autophagy receptor to mediate vacuolar targeting of soluble cargos. Mol. Cell 59, 1035–1042 (2015).
Oku, M. et al. Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 216, 3263–3274 (2017).
Mukherjee, A., Patel, B., Koga, H., Cuervo, A. M. & Jenny, A. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 12, 1984–1999 (2016).
Dice, J. F. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biol. Sci. 15, 305–309 (1990).
Cuervo, A. M. & Dice, J. F. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501–503 (1996).
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004).
Tsukamoto, S. et al. Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120 (2008).
Lim, J. & Yue, Z. Neuronal aggregates: Formation, clearance, and spreading. Dev. Cell 32, 491–501 (2015).
Komatsu, M. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006).
Hara, T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006).
Anding, A. L. & Baehrecke, E. H. Cleaning house: selective autophagy of organelles. Dev. Cell 41, 10–22 (2017).
Randow, F. & Youle, R. J. Self and nonself: How autophagy targets mitochondria and bacteria. Cell Host Microbe 15, 403–411 (2014).
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Nguyen, T. N., Padman, B. S. & Lazarou, M. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol. 26, 733–744 (2016).
McWilliams, T. G. et al. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439–449 (2018).
Haack, T. B. et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 91, 1144–1149 (2012).
Saitsu, H. et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat. Genet. 45, 445–449 (2013).
Kim, M. et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife 5, e12245 (2016).
Menzies, F. M. et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034 (2017).
Melendez, A. et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 (2003).
Madeo, F., Tavernarakis, N. & Kroemer, G. Can autophagy promote longevity? Nat. Cell Biol. 12, 842–846 (2010).
Lopez-Otin, C., Galluzzi, L., Freije, J. M., Madeo, F. & Kroemer, G. Metabolic control of longevity. Cell 166, 802–821 (2016).
Takamura, A. et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800 (2011).
Amaravadi, R., Kimmelman, A. C. & White, E. Recent insights into the function of autophagy in cancer. Genes Dev. 30, 1913–1930 (2016).
Zachari, M. & Ganley, I. G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 61, 585–596 (2017).
Noda, T. Autophagy in the context of the cellular membrane-trafficking system: the enigma of Atg9 vesicles. Biochem. Soc. Trans. 45, 1323–1331 (2017).
Kishi-Itakura, C., Koyama-Honda, I., Itakura, E. & Mizushima, N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127, 4089–4102 (2014).
Noda, T., Fujita, N. & Yoshimori, T. The late stages of autophagy: how does the end begin? Cell Death Differ. 16, 984–990 (2009).
Tsuboyama, K. et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041 (2016).
Nguyen, T. N. et al. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 215, 857–874 (2016).
Dunn, W. A. Jr. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 110, 1923–1933 (1990).
Axe, E. L. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 (2008).
Uemura, T. et al. A cluster of thin tubular structures mediates transformation of the ER to autophagic isolation membrane. Mol. Cell. Biol. 34, 1695–1706 (2014).
Hayashi-Nishino, M. et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 (2009).
Yla-Anttila, P., Vihinen, H., Jokitalo, E. & Eskelinen, E. L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185 (2009).
Graef, M., Friedman, J. R., Graham, C., Babu, M. & Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 24, 2918–2931 (2013).
Ge, L., Melville, D., Zhang, M. & Schekman, R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife 2, e00947 (2013).
Hamasaki, M. et al. Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013).
Nascimbeni, A. C. et al. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 36, 2018–2033 (2017).
Nishimura, T. et al. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 36, 1719–1735 (2017).
Noda, N. N., Ohsumi, Y. & Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 581, 1379–1385 (2010).
Rogov, V. V. et al. Structural and functional analysis of the GABARAP interaction motif (GIM). EMBO Rep. 18, 1382–1396 (2017).
Kamber, R. A., Shoemaker, C. J. & Denic, V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol. Cell 59, 372–381 (2015).
Lazarou, M. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 (2015).
Smith, M. D. et al. CCPG1is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell 44, 217–232 (2018).
Itakura, E. & Mizushima, N. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 192, 17–27 (2011).
Itakura, E., Kishi-Itakura, C., Koyama-Honda, I. & Mizushima, N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125, 1488–1499 (2012).
Li, Y., Lipowsky, R. & Dimova, R. Transition from complete to partial wetting within membrane compartments. J. Am. Chem. Soc. 130, 12252–12253 (2008).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Zhang, Y. et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136, 308–321 (2009).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Watanabe, T. et al. Genetic visualization of protein interactions harnessing liquid phase transitions. Sci. Rep. 7, 46380 (2017).
Zaffagnini, G. et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37, e98308 (2018).
Sun, D., Wu, R., Zheng, J., Li, P. & Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. http://doi.org/cmpd (2018).
Nguyen, N., Shteyn, V. & Melia, T. J. Sensing membrane curvature in macroautophagy. J. Mol. Biol. 429, 457–472 (2017).
Mi, N. et al. CapZ regulates autophagosomal membrane shaping by promoting actin assembly inside the isolation membrane. Nat. Cell Biol. 17, 1112–1123 (2015).
Seifert, U., Berndl, K. & Lipowsky, R. Shape transformations of vesicles: phase diagram for spontaneous-curvature and bilayer-coupling models. Phys. Rev. A. 44, 1182–1202 (1991).
Knorr, R. L., Dimova, R. & Lipowsky, R. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS ONE 7, e32753 (2012).
Campelo, F. et al. Sphingomyelin metabolism controls the shape and function of the Golgi cisternae. eLife 6, e24603 (2017).
Knorr, R. L., Lipowsky, R. & Dimova, R. Autophagosome closure requires membrane scission. Autophagy 11, 2134–2137 (2015).
Yu, S. & Melia, T. J. The coordination of membrane fission and fusion at the end of autophagosome maturation. Curr. Opin. Cell Biol. 47, 92–98 (2017).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Kuma, A., Komatsu, M. & Mizushima, N. Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619–1628 (2017).
Lu, Q. et al. Homeostatic control of innate lung inflammation by Vici syndrome gene Epg5 and additional autophagy genes promotes influenza pathogenesis. Cell Host Microbe 19, 102–113 (2016).
Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T. & Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18, 1042–1052 (2011).
Yamashita, S. I. et al. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 215, 649–665 (2016).
Nishida, Y. et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 (2009).
Malhotra, V. Unconventional protein secretion: an evolving mechanism. EMBO J. 32, 1660–1664 (2013).
Ponpuak, M. et al. Secretory autophagy. Curr. Opin. Cell Biol. 35, 106–116 (2015).
Green, D. R. & Levine, B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157, 65–75 (2014).
Cadwell, K. & Debnath, J. Beyond self-eating: the control of nonautophagic functions and signaling pathways by autophagy-related proteins. J. Cell Biol. 217, 813–822 (2018).
Bestebroer, J., V’Kovski, P., Mauthe, M. & Reggiori, F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic 14, 1029–1041 (2013).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016).
Levine, B., Packer, M. & Codogno, P. Development of autophagy inducers in clinical medicine. J. Clin. Invest. 125, 14–24 (2015).
Morel, E. et al. Autophagy: a druggable process. Annu. Rev. Pharmacol. Toxicol. 57, 375–398 (2017).
Mortimore, G. E. & Schworer, C. M. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 270, 174–176 (1977).
Pfeifer, U. Inhibition by insulin of the formation of autophagic vacuoles in rat liver: a morphometric approach to the kinetics of intracellular degradation by autophagy. J. Cell Biol. 78, 152–167 (1978).
Seglen, P. O. & Gordon, P. B. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA 79, 1889–1892 (1982).
Takeshige, K., Baba, M., Tsuboi, S., Noda, T. & Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 (1992).
Klionsky, D. J., Cueva, R. & Yaver, D. S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119, 287–299 (1992).
Blommaart, E. F., Luiken, J. J., Blommaart, P. J., van Woerkom, G. M. & Meijer, A. J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326 (1995).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Liang, X. H. et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 (1999).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Kihara, A., Noda, T., Ishihara, N. & Ohsumi, Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 (2001).
Ravikumar, B., Duden, R. & Rubinsztein, D. C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet .11, 1107–1117 (2002).
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004).
Bjørkøy, G. et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 (2005).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Rioux, J. D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Settembre, C. et al. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011).
Cullup, T. et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat. Genet. 45, 83–87 (2013).
Acknowledgements
I thank Willa Yim for expert editing of this manuscript, Roland Knorr for stimulating discussions on biomolecular condensates and physical models, and all members of my laboratory for helpful suggestions. This work was supported by the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research on Innovative Areas (Grant Numbers 25111001 and 25111005) and JST ERATO (Grant Numbers JPMJER1702).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol 20, 521–527 (2018). https://doi.org/10.1038/s41556-018-0092-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0092-5
This article is cited by
-
Mechanisms and functions of protein S-acylation
Nature Reviews Molecular Cell Biology (2024)
-
Autophagy-enhancing ATG16L1 polymorphism is associated with improved clinical outcome and T-cell immunity in chronic HIV-1 infection
Nature Communications (2024)
-
Comparison of different methods for preparation of nanochitosan as anticancer agent
Polymer Bulletin (2024)
-
Unraveling the role of autophagy regulation in Crohn's disease: from genetic mechanisms to potential therapeutics
Advanced Biotechnology (2024)
-
The Multiple Roles of Autophagy in Neural Function and Diseases
Neuroscience Bulletin (2024)