Abstract
Organ morphogenesis is a complex process coordinated by cell specification, epithelial–mesenchymal interactions and tissue polarity. A striking example is the pattern of regularly spaced, globally aligned mammalian hair follicles, which emerges through epidermal-dermal signaling and planar polarized morphogenesis. Here, using live-imaging, we discover that developing hair follicles polarize through dramatic cell rearrangements organized in a counter-rotational pattern of cell flows. Upon hair placode induction, Shh signaling specifies a radial pattern of progenitor fates that, together with planar cell polarity, induce counter-rotational rearrangements through myosin and ROCK-dependent polarized neighbour exchanges. Importantly, these cell rearrangements also establish cell fate asymmetry by repositioning radial progenitors along the anterior–posterior axis. These movements concurrently displace associated mesenchymal cells, which then signal asymmetrically to maintain polarized cell fates. Our results demonstrate how spatial patterning and tissue polarity generate an unexpected collective cell behaviour that in turn, establishes both morphological and cell fate asymmetry.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Biggs, L. C. & Mikkola, M. L. Early inductive events in ectodermal appendage morphogenesis. Semin. Cell Dev. Biol. 25–26, 11–21 (2014).
Ochoa-Espinosa, A. & Affolter, M. Branching morphogenesis: from cells to organs and back. Cold Spring Harb. Persp. Biol. 4, a008243 (2012).
St Johnston, D. & Ahringer, J. Cell polarity in eggs and epithelia: parallels and diversity. Cell 141, 757–774 (2010).
Campanale, J. P., Sun, T. Y. & Montell, D. J. Development and dynamics of cell polarity at a glance. J. Cell Sci. 130, 1201–1207 (2017).
Devenport, D. Tissue morphodynamics: translating planar polarity cues into polarized cell behaviors. Sem. Cell Dev. Biol. 55, 99–110 (2016).
Goodrich, L. V. & Strutt, D. Principles of planar polarity in animal development. Development 138, 1877–1892 (2011).
Yang, Y. & Mlodzik, M. Wnt-frizzled/planar cell polarity signaling: cellular orientation by facing the wind (Wnt). Annu. Rev. Cell. Dev. Biol. 31, 623–646 (2015).
Devenport, D. The cell biology of planar cell polarity. J. Cell Biol. 207, 171–179 (2014).
Butler, M. T. & Wallingford, J. B. Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 18, 375–388 (2017).
Adler, P. N. The frizzled/stan pathway and planar cell polarity in the Drosophila wing. Curr. Top. Dev. Biol. 101, 1–31 (2012).
Sennett, R. & Rendl, M. Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin. Cell Dev. Biol. 23, 917–927 (2012).
Duverger, O. & Morasso, M. I. Epidermal patterning and induction of different hair types during mouse embryonic development. Birth Defects Res. C 87, 263–272 (2009).
Millar, S. E. Molecular mechanisms regulating hair follicle development. J. Invest Dermatol. 118, 216–225 (2002).
Devenport, D. & Fuchs, E. Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat. Cell Biol. 10, 1257–1268 (2008).
Wang, Y., Chang, H. & Nathans, J. When whorls collide: the development of hair patterns in frizzled 6 mutant mice. Development 137, 4091–4099 (2010).
Levy, V., Lindon, C., Harfe, B. D. & Morgan, B. A. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev. Cell 9, 855–861 (2005).
Harfe, B. D. et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Aigouy, B., Umetsu, D. & Eaton, S. Segmentation and quantitative analysis of epithelial tissues. Methods Mol. Biol. 1478, 227–239 (2016).
Guillot, C. & Lecuit, T. Mechanics of epithelial tissue homeostasis and morphogenesis. Science 340, 1185–1189 (2013).
Zallen, J. A. & Blankenship, J. T. Multicellular dynamics during epithelial elongation. Semin. Cell Dev. Biol. 19, 263–270 (2008).
Aw, W. Y., Heck, B. W., Joyce, B. & Devenport, D. Transient tissue-scale deformation coordinates alignment of planar cell polarity junctions in the mammalian skin. Curr. Biol. 26, 2090–2100 (2016).
Cetera, M., Leybova, L., Woo, F. W., Deans, M. & Devenport, D. Planar cell polarity-dependent and independent functions in the emergence of tissue-scale hair follicle patterns. Dev. Biol. 428, 188–203 (2017).
Chang, H., Smallwood, P. M., Williams, J. & Nathans, J. The spatio-temporal domains of Frizzled6 action in planar polarity control of hair follicle orientation. Dev. Biol. 409, 181–193 (2016).
Bertet, C., Sulak, L. & Lecuit, T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667–671 (2004).
Blankenship, J. T., Backovic, S. T., Sanny, J. S., Weitz, O. & Zallen, J. A. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell 11, 459–470 (2006).
Nishimura, T., Honda, H. & Takeichi, M. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149, 1084–1097 (2012).
Williams, M., Yen, W., Lu, X. & Sutherland, A. Distinct apical and basolateral mechanisms drive planar cell polarity-dependent convergent extension of the mouse neural plate. Dev. Cell 29, 34–46 (2014).
Shindo, A. & Wallingford, J. B. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science 343, 649–652 (2014).
Jamora, C., DasGupta, R., Kocieniewski, P. & Fuchs, E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422, 317–322 (2003).
Bitgood, M. J. & McMahon, A. P. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev. Biol. 172, 126–138 (1995).
Nowak, J. A., Polak, L., Pasolli, H. A. & Fuchs, E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell 3, 33–43 (2008).
Ouspenskaia, T., Matos, I., Mertz, A. F., Fiore, V. F. & Fuchs, E. WNT-SHH antagonism specifies and expands stem cells prior to niche formation. Cell 164, 156–169 (2016).
Xu, Z. et al. Embryonic attenuated Wnt/beta-catenin signaling defines niche location and long-term stem cell fate in hair follicle. eLife 4, e10567 (2015).
Vidal, V. P. et al. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol. 15, 1340–1351 (2005).
St-Jacques, B. et al. Sonic hedgehog signaling is essential for hair development. Curr. Biol. 8, 1058–1068 (1998).
Karlsson, L., Bondjers, C. & Betsholtz, C. Roles for PDGF-A and sonic hedgehog in development of mesenchymal components of the hair follicle. Development 126, 2611–2621 (1999).
Chiang, C. et al. Essential role for Sonic hedgehog during hair follicle morphogenesis. Dev. Biol. 205, 1–9 (1999).
Mill, P. et al. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 17, 282–294 (2003).
Schweisguth, F. Asymmetric cell division in the Drosophila bristle lineage: from the polarization of sensory organ precursor cells to Notch-mediated binary fate decision. WIRES Dev. Biol. 4, 299–309 (2015).
Peng, Y., Han, C. & Axelrod, J. D. Planar polarized protrusions break the symmetry of EGFR signaling during Drosophila bract cell fate induction. Dev. Cell 23, 507–518 (2012).
Capilla, A. et al. Planar cell polarity controls directional Notch signaling in the Drosophila leg. Development 139, 2584–2593 (2012).
Cooper, M. T. & Bray, S. J. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397, 526–530 (1999).
Fanto, M. & Mlodzik, M. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature 397, 523–526 (1999).
Tomlinson, A. & Struhl, G. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development 126, 5725–5738 (1999).
Driskell, R. R., Clavel, C., Rendl, M. & Watt, F. M. Hair follicle dermal papilla cells at a glance. J. Cell Sci. 124, 1179–1182 (2011).
Ahtiainen, L. et al. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev. Cell 28, 588–602 (2014).
Glover, J. D. et al. Hierarchical patterning modes orchestrate hair follicle morphogenesis. PLoS Biol. 15, e2002117 (2017).
Mlodzik, M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. EMBO J. 18, 6873–6879 (1999).
Keller, R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science 298, 1950–1954 (2002).
Tada, M. & Heisenberg, C. P. Convergent extension: using collective cell migration and cell intercalation to shape embryos. Development 139, 3897–3904 (2012).
Shindo, A. Models of convergent extension during morphogenesis. WIRES Dev. Biol. 7, e293 (2017).
Zheng, L., Zhang, J. & Carthew, R. W. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development 121, 3045–3055 (1995).
Theisen, H. et al. dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development 120, 347–360 (1994).
Wolff, T. & Rubin, G. M. Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development 125, 1149–1159 (1998).
Winter, C. G. et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105, 81–91 (2001).
Fiehler, R. W. & Wolff, T. Drosophila myosin II zipper is essential for ommatidial rotation. Dev. Biol. 310, 348–362 (2007).
Chuai, M. & Weijer, C. J. The mechanisms underlying primitive streak formation in the chick embryo. Curr. Top. Dev. Biol. 81, 135–156 (2008).
Rozbicki, E. et al. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat. Cell Biol. 17, 397–408 (2015).
Voiculescu, O., Bertocchini, F., Wolpert, L., Keller, R. E. & Stern, C. D. The amniote primitive streak is defined by epithelial cell intercalation before gastrulation. Nature 449, 1049–1052 (2007).
Aigouy, B. et al. Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell 142, 773–786 (2010).
Acknowledgements
We gratefully acknowledge those who provide mouse lines, technical support, and valuable discussions that contributed to this project. We thank Saori Haigo and Jeremy Reiter for generous donation of Vangl2 and Fz6 alleles in mT/mG backgrounds. Michael Deans and Jeremy Nathans kindly provided the Vangl2 and Vangl1 floxed mouse lines. Beniot Aiguoy developed and distributed Packing Analyzer v2 and Tissue Analyzer software for image analysis. We thank Katie Little for assistance with genetic crosses and genotyping, and members of the Devenport lab for insightful comments and suggestions. Finally, we thank Gary Laevsky for imaging support and expertise. The Confocal Facility at Princeton University is a Nikon Center of Excellence. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number R01AR066070 and a Vallee Foundation Scholars Award to D.D. L.L. was supported by NIH training grant T32 GM007388.
Author information
Authors and Affiliations
Contributions
Conceptualization, M.C., B.J., and D.D.; Investigation, M.C., L.L., B.J., and D.D..; Writing, M.C., L.L and D.D.; Funding Acquisition and Supervision, D.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
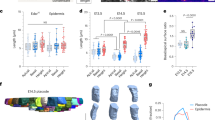
Supplementary Figure 1 Cell movements during placode polarization.
(A-D) Spinning disk confocal images from a time series showing placode polarization. Representative placodes from three embryos displaying similar cell movements shown in Figure 1D. Cells express mTomato (magenta) and mGFP driven by K14Cre (green). Mosaicsm allows tracking of small clones through time. See Supplemental Video 3. Coloured dots indicate the same cells through time. The z plane changes in 3μm steps to follow the base of the placode into the dermis. (A) Cells that are initially located at the center of the placode are displaced anteriorly. Supplemental Video 3, left. (B) Cells at the posterior of the placode converge toward the placode midline. Supplemental Video 3, center. (C) Cells at the anterior edge of the placode are displaced laterally and lateral cells are displaced posteriorly. Supplemental Video 3, right. (D) Groups of cells located laterally rotate as cells closest to the center of the placode move anteriorly and cells at the lateral edge move posteriorly. The same placode is used in panels B and D with different groups of cells highlighted. Supplemental Video 3, center. Scale bar, 10μm. Anterior is to the left.
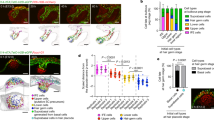
Supplementary Figure 2 Celsr1 polarity in early and polarizing placodes.
Additional examples of Celsr1 asymmetry in early (left) and polarizing placodes (right) representative of 15 measured images from four embryos shown in Figure 2C. The orientation of the line shows the direction of Celsr polarity relative to the AP axis (0 degrees). 0–44 degrees is shown in cyan and 45–90 degrees is shown in yellow. Scale bar, 10μm. Anterior is to the left.
Supplementary Figure 3 Counter-rotational cell flows require planar cell polarity.
Additional example of Fz6 KO placode polarization shown in Figure 3B representative from three embryos. In this example, Fz6 KO cells undergo reduced counter-rotational cell movements, which correlate with the direction of placode growth rather than the AP axis. Spinning disk confocal images from a time series of placode cells expressing mTomato. Cells were segmented and false coloured in a rainbow pattern of vertical lines perpendicular to the direction of growth at the start of the video. Cell tracks show the movement of cells during the designated time window (bottom). Overall cell trajectories are shown in the schematic (right). The z plane changes 3μm in one step to follow the base of the placode into the dermis. Scale bar, 10μm. Anterior is to the left.
Supplementary Figure 4 Placode polarization and counter rotational movements require myosin II and Rho kinase activity.
(A-C) Representative immunofluorescence images of explants cultured from E15.5 in the presence of DMSO (A), blebbistatin (B), and Y-27632 (C) quantified in D. Follicles are labeled with Shh-Cre driving mGFP expression. 1st, 2nd, and 3rd wave follicles are present under all conditions. 1st wave follicles remain polarized in the presence of blebbistatin and Y-27632. Scale bar, 50 µm. (D) Quantification of hair follicle frequencies at each stage after 24 hours in culture. Control, n=649 follicles from 7 embryos and blebbistatin, n=611 follicles from 7 embryos (mean+SD. p= n.s. for all waves control vs blebbistatin, unpaired t-test); control, n=271 follicles from 4 embryos and Y-27632, n=244 follicles from 4 embryos (mean+SD. p= n.s. for all waves control vs. Y-27632). (E) Additional representative placode shown in Figure 4E from five treated explants. Y-27632 inhibits counter-rotational cell flows. Spinning disk confocal images from a 13.3 hour time series of placode cells expressing mTomato (top). Cells were segmented and false coloured in a rainbow pattern of vertical lines perpendicular to the AP axis at the beginning of the video. Cell tracks show the movement of cells during the designated time window (bottom). Overall cell trajectories are shown in the schematic (right). Cells remain in their original rainbow pattern. Scale bar, 10μm. Anterior is to the left.
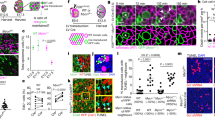
Supplementary Figure 5 Characterization of placode morphology and cell movements in Shh mutants.
(A-C) Shh KO embryos display expanded and irregular shaped placodes. (A) In controls, Shh-GFP (green) and P-Cadherin (magenta) mark the inner placode. In Shh KO embryos, the zone of Shh-GFP and P-Cadherin is expanded, irregularly shaped, and lacks a smooth boundary between GFP+ and GFP- cells. Representative mild and severe Shh KO examples along with a control, quantified in B. (B) Quantification of placode morphologies. The area and perimeter of Shh KO placodes, as measured by the zone of Shh-GFP expression, are expanded compared to controls. Shh KO placodes are less circular and more anisotropic than controls. n=30 control placodes from 3 embryos; n= 40 ShhKO placodes from 4 embryos. Lines show mean+SD. p=1.25x10−16 for area, p=1.06x10−16 for perimeter, p= 8.82x10−7 for circularity, p= 8.78x10−7 for aspect ratio, two-tailed t-tests with Welch’s correction for unequal variances (except for aspect ratio, where variances are not significantly different). (C) Shh-Cre>mGFP highlights the irregular morphology and poor compartmentalization of Shh KO placodes. E15.5 explants were cultured overnight before fixation. Representative images from two control and two mutant embryos. (D) Shh KO placode cells undergo atypical cell rearrangements. Additional example to Figure 5C. Spinning disk confocal images from a time series of placode cells expressing mTomato (top). Cells were segmented and false coloured in a rainbow pattern of vertical lines perpendicular to the AP axis at the beginning of the video. Cell tracks show the movement of cells during the designated time window (bottom). Additional placode representative of two embryos. Overall cell trajectories are shown in the schematic (right). Scale bar, 10μm. Anterior is to the left.
Supplementary Figure 6 Cell fate asymmetry requires myosin II and Rho kinase activity.
Shh-Cre driving mGFP marks anterior placode cell fates (green) while Sox9 marks posterior placode cell fates (magenta). Representative immunofluorescence images of germ placodes cultured from E15.5 in the presence of DMSO (left), blebbistatin (center), and Y-27632 (right) quantified in Figure S4D. Shh and Sox9 expressing cells fail to separate along the AP axis in the presence of myosin or Rho kinase inhibitors. Scale bar, 10µm. Anterior is to the left.
Supplementary information
Supplementary Information
Supplementary Figures 1–6, Supplementary Table and Video legends
Supplementary Table 1
Mouse genotypes.
Supplementary Table 2
Statistics source data.
Supplementary Video 1
Live imaging of embryonic skin development.
Supplementary Video 2
Live imaging of hair placode polarization.
Supplementary Video 3
Region specific cell movements during placode polarization.
Supplementary Video 4
Live imaging of tracked cells during placode polarization shows counter-rotational cell movements.
Supplementary Video 5
Counter-rotational cell movements fail to occur in Vangl mutants.
Supplementary Video 6
Counter-rotational cell movements occur in the direction of placode growth in Fz6 mutants.
Supplementary Video 7
Rho kinase inhibitor blocks counter-rotational cell movements in mid-polarizing placodes.
Supplementary Video 8
Shh mutant placodes undergo extensive cell rearrangements.
Supplementary Video 9
Counter-rotational cell flows within the placode polarize the dermal condensate.
Rights and permissions
About this article
Cite this article
Cetera, M., Leybova, L., Joyce, B. et al. Counter-rotational cell flows drive morphological and cell fate asymmetries in mammalian hair follicles. Nat Cell Biol 20, 541–552 (2018). https://doi.org/10.1038/s41556-018-0082-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0082-7
This article is cited by
-
Mechanical forces across compartments coordinate cell shape and fate transitions to generate tissue architecture
Nature Cell Biology (2024)
-
Anillin governs mitotic rounding during early epidermal development
BMC Biology (2022)
-
Exploring the Potential of Mesenchymal Stem Cell–Derived Exosomes for the Treatment of Alopecia
Regenerative Engineering and Translational Medicine (2021)
-
The role of single-cell mechanical behaviour and polarity in driving collective cell migration
Nature Physics (2020)