Abstract
The contribution of autophagy to cancer development remains controversial, largely owing to the fact that autophagy can be tumour suppressive or oncogenic in different biological contexts. Here, we show that in non-small-cell lung cancer (NSCLC), casein kinase 1 alpha 1 (CK1α) suppresses tumour growth by functioning as an autophagy inducer to activate an autophagy-regulating, tumour-suppressive PTEN/AKT/FOXO3a/Atg7 axis. Specifically, CK1α bound the C-terminal tail of PTEN and enhanced both PTEN stability and activity by competitively antagonizing NEDD4-1-induced PTEN polyubiquitination and abrogating PTEN phosphorylation, thereby inhibiting AKT activity and activating FOXO3a-induced transcription of Atg7. Notably, blocking CK1α-induced Atg7-dependent autophagy cooperates with oncogenic HRasV12 to initiate tumorigenesis of lung epithelial cells. An association of a CK1α-modulated autophagic program with the anti-neoplastic activities of the CK1α/PTEN/FOXO3a/Atg7 axis was demonstrated in xenografted tumour models and human NSCLC specimens. This provides insights into the biological and potentially clinical significance of autophagy in NSCLC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Boya, P., Reggiori, F. & Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 15, 713–720 (2013).
Elgendy, M., Sheridan, C., Brumatti, G. & Martin, S. J. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol. Cell 42, 23–35 (2011).
Kitanaka, C. et al. Increased Ras expression and caspase-independent neuroblastoma cell death: possible mechanism of spontaneous neuroblastoma regression. J. Natl Cancer Inst. 94, 358–368 (2002).
Marino, G., Martins, I. & Kroemer, G. Autophagy in Ras-induced malignant transformation: fatal or vital? Mol. Cell 42, 1–3 (2011).
White, E. The role for autophagy in cancer. J. Clin. Invest. 125, 42–46 (2015).
White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 12, 401–410 (2012).
Mizushima, N. Autophagy: process and function. Genes Dev. 21, 2861–2873 (2007).
Klionsky, D. J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937 (2007).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu Rev. Cell Dev. Biol. 27, 107–132 (2011).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Ohsumi, Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216 (2001).
Strohecker, A. M. et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 3, 1272–1285 (2013).
Wu, Y. T., Tan, H. L., Huang, Q., Ong, C. N. & Shen, H. M. Activation of the PI3K-Akt-mTOR signaling pathway promotes necrotic cell death via suppression of autophagy. Autophagy 5, 824–834 (2009).
Wang, R. C. et al. Akt-mediated regulation of autophagy and tumorigenesis through beclin 1 phosphorylation. Science 338, 956–959 (2012).
Dansen, T. B. & Burgering, B. M. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18, 421–429 (2008).
Zhao, Y. et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 12, 665–688 (2010).
Arico, S. et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 276, 35243–35246 (2001).
Gozuacik, D. & Kimchi, A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23, 2891–2906 (2004).
Yanagi, S. et al. Pten controls lung morphogenesis, bronchioalveolar stem cells, and onset of lung adenocarcinomas in mice. J. Clin. Invest. 117, 2929–2940 (2007).
Chalhoub, N. & Baker, S. J. PTEN and the PI3-kinase pathway in cancer. Annu Rev. Pathol. 4, 127–150 (2009).
Marsit, C. J. et al. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum. Pathol. 36, 768–776 (2005).
Wang, X. J. et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 (2007).
Amodio, N. et al. Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a PTEN negative regulator, in non-small-cell lung carcinomas. Am. J. Pathol. 177, 2622–2634 (2010).
Maddika, S. et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–U224 (2011).
Gericke, A., Munson, M. & Ross, A. H. Regulation of the PTEN phosphatase. Gene 374, 1–9 (2006).
Odriozola, L., Singh, G., Hoang, T. & Chan, A. M. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J. Biol. Chem. 282, 23306–23315 (2007).
Sinnberg, T. et al. Suppression of casein kinase 1α in melanoma cells induces a switch in β-catenin signaling to promote metastasis. Cancer Res. 70, 6999–7009 (2010).
Pribluda, A. et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell 24, 242–256 (2013).
Elyada, E. et al. CKIα ablation highlights a critical role for p53 in invasiveness control. Nature 470, 409–413 (2011).
Al-Khouri, A. M., Ma, Y., Togo, S. H., Williams, S. & Mustelin, T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3β. J. Biol. Chem. 280, 35195–35202 (2005).
Mammucari, C. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007).
Chaanine, A. H. et al. JNK modulates FOXO3a for the expression of the mitochondrial death and mitophagy marker BNIP3 in pathological hypertrophy and in heart failure. Cell Death Dis. 3, 265 (2012).
Xu, P., Das, M., Reilly, J. & Davis, R. J. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 25, 310–322 (2011).
Young, A. R. et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 23, 798–803 (2009).
Patel, K. K. & Stappenbeck, T. S. Autophagy and intestinal homeostasis. Annu Rev. Physiol. 75, 241–262 (2013).
Cheong, J. K. et al. Casein kinase 1α-dependent feedback loop controls autophagy in RAS-driven cancers. J. Clin. Invest. 125, 1401–1418 (2015).
Szyniarowski, P. et al. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 7, 892–903 (2011).
Schneider, R. K. et al. Role of casein kinase 1A1 in the biology and targeted therapy of del(5q) MDS. Cancer Cell 26, 509–520 (2014).
Kollmann, K. et al. A kinase-independent function of CDK6 links the cell cycle to tumor angiogenesis. Cancer Cell 24, 167–181 (2013).
Rao, S. et al. A dual role for autophagy in a murine model of lung cancer. Nat. Commun. 5, 3056 (2014).
Song, M. S., Salmena, L. & Pandolfi, P. P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296 (2012).
Vazquez, F., Ramaswamy, S., Nakamura, N. & Sellers, W. R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell Biol. 20, 5010–5018 (2000).
Vazquez, F. et al. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 276, 48627–48630 (2001).
Georgescu, M. M., Kirsch, K. H., Akagi, T., Shishido, T. & Hanafusa, H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc. Natl Acad. Sci. USA 96, 10182–10187 (1999).
Wang, X. J., Shi, Y. J., Wang, J. R., Huang, G. C. & Jiang, X. J. Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. Biochem. J. 414, 221–229 (2008).
Yim, E. K. et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell 15, 304–314 (2009).
Yim, E. K., Siwko, S. & Lin, S. Y. Exploring Rak tyrosine kinase function in breast cancer. Cell Cycle 8, 2360–2364 (2009).
Cance, W. G. et al. Rak, a novel nuclear tyrosine kinase expressed in epithelial cells. Cell Growth Differ. 5, 1347–1355 (1994).
Park, S. M. et al. U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3’ end formation. Mol. Cell 62, 479–490 (2016).
Takamura, A. et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800 (2011).
Guo, J. Y. et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Gene Dev. 27, 1447–1461 (2013).
Karsli-Uzunbas, G. et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4, 914–927 (2014).
Chen, S. & Guan, J. L. Tumor-promoting and -suppressive roles of autophagy in the same mouse model of BrafV600E-driven lung cancer. Cancer Discov. 3, 1225–1227 (2013).
Levine, B., Mizushima, N. & Virgin, H. W. Autophagy in immunity and inflammation. Nature 469, 323–335 (2011).
Gazdar, A. F. & Oie, H. K. Cell culture methods for human lung cancer. Cancer Genet. Cytogenet. 19, 5–10 (1986).
Oie, H. K., Russell, E. K., Carney, D. N. & Gazdar, A. F. Cell culture methods for the establishment of the NCI series of lung cancer cell lines. J. Cell Biochem. 24, 24–31 (1996).
Cai, J. et al. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J. Clin. Invest. 123, 566–579 (2013).
Tran, H. et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45. Protein Sci. 296, 530–534 (2002).
Wang, H. W. et al. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat. Genet. 36, 687–693 (2004).
Maddika, S. & Chen, J. J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat. Cell Biol. 11, 409–110 (2009).
Shenoy, S. et al. Membrane association of the PTEN tumor suppressor: molecular details of the protein-membrane complex from SPR binding studies and neutron reflection. PLoS ONE 7, e32591 (2012).
Acknowledgements
This work was supported by Key Program of the National Natural Science Foundation of China (81330058); the National Key Research and Development Program of China (2017YFA0505600, 2017YFA0106300); the Natural Science Foundation of China (81621004, 81472574, 81772473, 81472351); China’s Marine Commonwealth Research Project (201305017); the Natural Science Foundation of Guangdong Province (2016A030306026, 2015A030313138); Pearl River S&T Nova Program of Guangzhou (201610010177); Guandong Science and Technology Project (2015B090903063);. the Fundamental Research Funds for the Central Universities (17ykzd06).
Author information
Authors and Affiliations
Contributions
Project planning was done by M.L. and J.C.; J.C., R.L. and X.X. performed the experiments, analysed data and wrote a draft of the paper with the help of L.F., Y.H., L.Z., R.Lian, X.F., X.L., X.Li and J.W. X.Z., H.Z. and F.H. provided the associated clinical data from the subjects and provided technical assistance and expertise in the lung tumour sample preparation. J.C. and R.L. contributed to the statistical analyses of the data. J.L., M.Z., E.S., Y.He and Y.Y. discussed experiments and edited the manuscript. M.L. conceived the ideas, designed and discussed experiments, supervised progress and extensively edited and communicated regarding the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
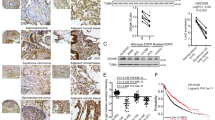
Supplementary Figure 1. CK1α induces potent cellular autophagy and inhibits NSCLC cell growth.
(a) Western blotting (WB) or qRT-PCR analysis of CK1α expression and LC3 conversion in A549 NSCLC cells in response to autophagy inducer EBSS, rapamycin, brefeldin A or LiCl, or autophagy inhibitor 3-MA. (b and c) Representative images and the percentages of cells showing GFP-LC3 puncta accumulation in indicated cells with enforced or silenced CK1α expression in the absence or presence of treatment with autophagy inducer EBSS. (d) WB analysis of changes induced by CK1α overexpression or knockdown in LC3 conversion, Atg7 and p62 levels in 293FT cells in the absence or presence of Baf-A1 treatment. (e and f) qRT-PCR or WB analysis of a panel of autophagy-related genes, including Atg4, Atg5, Atg7, Atg16 and LC3B in indicated cells. (g and h) Representative images and quantification of cell colonies of indicated cells grown in the 2-D colony formation assay (g) or in the 3-D soft-agar assay (h). Scale bar: 10 μm (b and c), 100 μm (h). All experiments were repeated three times with similar results and images/immunoblots in a-h are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01). Source data for precise P values are available in Supplementary Table 4.
Supplementary Figure 2 CK1α interacts with PTEN to abrogate PTEN phosphorylation and prolong PTEN half-life.
. (a) Immunoprecipitation assay revealing interaction between HA-PTEN with endogenous CK1α. (b) Immunoprecipitation assay revealing interaction between FALG-CK1α and indicated nonphosphorylatable (indicated residues mutated to alanine) and phosphomimic mutants (indicated residues mutated to aspartic acid) of HA-PTEN in 293FT cells. (c) Schematic representation of full-length CK1α (FL), along with various deletion mutants as indicated. The interactions between PTEN and CK1α were determined by immunoprecipitation assay in 293FT cells. (d) Intensities of immunoblotting bands were quantified, and the relative ratio of phosphorylated PTEN to total PTEN for 293FT cells is shown. (e) WB analysis of PTEN phosphorylation levels after ectopic HA-PTEN was immunoprecipitated in indicated cells treated with MG132. (f) The effect of overexpressing or silencing CK1α on PTEN phosphorylation. (g) Immunoprecipitated components, using anti-FALG affinity purification, from lysates of indicated cells transfected with FALG-CK1α were examined with the levels of phosphorylated PTEN at Thr366 or Ser385. (h) SPR analysis measuring the affinity and kinetics of the interaction between PTEN with GSK3β or CK2. PTEN was immobilized on a CM5 chip. (i) Immunoprecipitation assay revealing no binding of PTEN to GSK3β or CK2 in various NSCLC cells. (j) WB analysis of the effect of overexpressing or silencing GSK3β or CK2 on phosphorylation of PTEN. (k) Silencing CK1α promoted PTEN polyubiquitination in SLC cells. (l) SPR analysis measuring the affinity and kinetics of the interaction between CK1α and C-terminal tail residues of PTEN fused to the C-terminus of EGFP, wild-type EGFP or a PTEN mutant (ΔPTEN) with its C-terminus deleted, respectively. CK1α was immobilized on a CM5 chip. (m) WB analysis of the effect of a nonphosphorylatable mutant (S380A-T382A-T383A, 3S/T-3A), a phosphomimic mutant (T366D) of PTEN and the ΔPTEN on AKT phosphorylation in indicated cells. (n) The effect of overexpressing or silencing CK1α on protein half-lives of indicated PTEN mutants. All experiments were repeated three times with similar results and images/immunoblots in a-c and e-m are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01). Source data for precise P values are available in Supplementary Table 4.
Supplementary Figure 3 CK1α exerts pro-autophagic effect and tumor-suppressive effect independent of catalytic activity of the kinase.
. (a) In vitro phosphorylation assay revealing no direct phosphorylation of recombinant GST-PTEN by purified His-CK1α. (b) The percentages of cells showing GFP-LC3 puncta accumulation in indicated cells. (c) WB analysis of changes in LC3 conversion, Atg7 and p62 levels in indicated cells. (d) Quantification of cell colonies of indicated cells grown in the 3-D soft agar assay. (e) Indicated cells overexpressing indicated mutants of CK1α, and corresponding vectot-control cells, were subcutaneously transplanted in nude mice (n = 5 biologically independent mice per group). Subcutaneous tumors at the experimental endpoint and tumor growth curves are shown as indicated. (f) Tumor tissue samples taken from xenografts of indicated A549 cells were examined with ultrastructural cytoplasimic autophagosomes and/or autolysosomes by TEM observation. (g) WB analysis of changes in total PTEN, phospholation of PTEN at thr366, AKT at ser473 and FOXO3a at ser253 induced by overexpression of indicated mutants of CK1α. (h) Immunoprecipitation assay revealing binding between PTEN and indicated mutants of CK1α in indicated cells with indicated treatments. (i and j) The effect of CK1α inhibition by a small-molecule inhibitor D4476 or by shRNA silencing CK1α on the autophagic flux in the absence or presence of, respectively, 5μM CQ treatment. (k and l) A549 or SLC cells overexpressing or silenced for CK1α, and corresponding control cells, were subcutaneously transplanted in nude mice (n = 5 biologically independent mice per group). 15 mpk D4476, 15 mpk CQ, or 15 mpk:15 mpk D4476:CQ were injected i.p. daily. Subcutaneous tumors at the experimental endpoint and tumor growth curves are shown as indicated. Scale bar: 1 cm (e, k and l), and 200 nm (f). All experiments were repeated three times with similar results and images/immunoblots in a, c, e-h, and j-l are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01). Source data for precise P values are available in Supplementary Table 4.
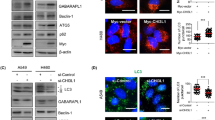
Supplementary Figure 4 PTEN is essential for CK1α-induced potent autophagy and suppression of tumor growth.
. (a) PTEN promoter methylation was detected by MSP, and PTEN expression was evaluated by immunoblotting in indicated NSCLC cell lines. (b) Immunoprecipitation assay reveals exogenous interaction between HA-PTEN with FLAG-CK1α in both PTEN-negative and PTEN-restored PLLC cells. (c) WB was performed to determine the effect of dose-dependently increasing or decreasing CK1α expression on PTEN protein levels and its phosphorylation, in both PTEN-negative and PTEN-restored PLLC cells. (d-g) PTEN-negative and PTEN-restored PLLC cells were enforced to express CK1α or the ΔCK1α mutant, or silenced for CK1α. Autophagy assays, including observation of LC3-immunopositive punctate signals (d), TEM observation (e), real-time quantification of deregulated expression of autophagy-related genes (f), and immunoblotting detection of changes in CK1α, LC3 conversion, Atg7 and p62 (g), were performed. Arrows in red indicate autophagosomes or autolysosomes. (h-j) The effects of overexpressing or CK1α in PTEN silenced H460 or SLC cells on autophagy and tumor growth were, separately, evaluated by measuring LC3-immunopositive punctate signals (h), WB analysis of changes in CK1α, LC3 conversion, Atg7 and p62 (i), and 3-D soft agar assay (j). (k and l) Indicated cells were subcutaneously transplanted in nude mice (n = 5 biologically independent mice per group). Tumor growth curves (k) and subcutaneous tumors at the experimental endpoint (l) were shown as indicated. (m) Tumor tissues from xenografts of indicated cells were examined with ultrastructural cytoplasimic autophagosomes and/or autolysosomes by TEM observation. Scale bar: 10 μm (d and h), 200 nm (e and m) and 1 cm (l). All experiments were repeated three times with similar results and images/immunoblots in a-i, l and m are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01, NS: not statistically significant). Source data for precise P values are available in Supplementary Table 4.
Supplementary Figure 5 AKT/FOXO3a signaling essentially mediates CK1α-induced autophagy and suppression of tumor growth.
. (a) Immunoblotting results show no impact of either increasing or decreasing CK1α expression on β-catenin phosphorylation at ser45, Beclin1 phosphorylation at ser234, or phosphorylation of S6K or 4EBP1 in indicated NSCLC cells. (b) TOP/FOP reporter assays show no impact of either increasing or decreasing CK1α expression on β-catenin-driven transcription. (c and d) The effects of inhibiting AKT activity with two distinct AKT inhibitors, namely, AKT-in-VIII and AKT-in-IX, on FOXO3a transcriptional activities (c) and expression of indicated autophagy-related genes (d). (e) The impact of silencing FOXO3a on the expression of autophagy related genes in indicated CK1α-overexpressing cells. (f) Representative images and quantification of cell colonies of indicated cells grown in soft agar are shown. PTEN-silenced SLC cells were treated with AKT-in-IX, and furthur silenced for FOXO3a (left panel). PTEN-restored PLLC cells were silenced with FOXO3a (middle panel) or transduced with a constitutively activated mutant of AKT (myr-AKT) followed by overexpression of a constitutively activated mutant of FOXO3a (S253A) or a Beclin1 mutant (S234A) or treatment with two distinct mTOR inhibitors (Rapamycin and AZD8055) separately (right panel). (g) WB analysis determining the effect of CK1α on FOXO3a phosphorylation at ser318/321 and ser253 and total protein levels of FOXO3a. (h) In vitro phosphorylation assay revealing no direct phosphorylation of recombinant GST-FOXO3a by purified His-CK1α. (i) Immunoprecipitation assay revealing no binding of FOXO3a with CK1α in indicated NSCLC cells. (j-l) The effect of separately overexpressing indicated mutants of FOXO3a or silencing FOXO3a in CK1α-silenced PLLC-PTEN cells on autophagy or growth ability in soft agar were evaluated. (m) Immunoprecipitation analysis of the interaction between CK1α to PTEN or FOXO3a in SW48 and LS123 cells carrying p53 hotpot mutations and expressing relatively high-level PTEN. (n) WB analysis of changes in LC3 conversion, Atg7, p62 and PTEN levels and phosphorylation levels of AKT and FOXO3a in indicated cells with indicated treatments. All experiments were repeated three times with similar results and immunoblots in a, g-i, k, m and n are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01). Source data for precise P values are available in Supplementary Table 4.
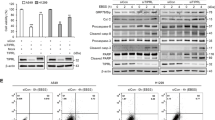
Supplementary Figure 6 Atg7 plays an important role in CK1α-induced autophagy.
. (a and b) WB assays were performed to determine the impact of overexpressing or silencing FOXO3a on protein expression, respectively, of Atg4, Atg5, LC3B and Atg16 in indicated cells. (c and d) WB analysis of basal LC3 conversion, protein levels of Atg4, Atg5, Atg16 and Atg7 in four pairs of human NSCLC tumor tissues and paired adjacent normal tissues freshly collected (c) and indicated cells (d). (e) Detection of the autophagic flux with the mRFP-GFP-LC3 reporter in CK1α-overexpressing PLLC-PTEN cells silenced for Atg7, in the absence or presence of Baf-A1 treatment. Scale bar: 5 μm (e). (f and g) WB analysis of total protein levels of Atg7, CK1α, PTEN and FOXO3a and phosphorylation of PTEN(Thr366), AKT(Ser473) and FOXO3a(Ser253), and qRT-PCR analysis of mRNA levels of CK1α, Atg7 and PTEN in 8 pairs of human NSCLC tumor tissues and paired adjacent normal tissues freshly collected (f). Correlation analysis of CK1α, PTEN, and Atg7 protein levels, and PTEN, AKT, and FOXO3a phosphorylation levels in these 8 pairs of freshly collected human NSCLC tumor tissues and paired adjacent normal tissues. Intensities of immunoblotting bands were quantified by scanning densitometry (g). n = 16 biologically independent samples. Pearson’s correlation test (two-sided) was used to compute the r (R2) values. (h) WB analyses of CK1α protein levels in two primary normal lung epithelial cells and a panel of NSCLC cell lines (n = 16 biologically independent samples). (i) The TCGA lung cancer dataset shows Atg7 mRNA levels in paired non-tumor tissues and tumor tissues from NSCLC patients (n = 71 biologically independent human participants). All experiments were repeated three times with similar results and images/immunoblots in a-g are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01). Source data for precise P values are available in Supplementary Table 4.
Supplementary Figure 7 The CK1α/PTEN/AKT/FOXO3a/Atg7 axis requires a distinct set of Atgs to induce autophagy and suppress NSCLC cell growth.
. (a-e) CK1α-overexpressing NSCLC cells and PTEN-restored PLLC cells in parallel with PTEN-negative PLLC cells were separately silenced with Atg5, LC3B, Atg7, Atg4 or Atg16, followed by immunoblotting detection of changes in LC3 conversion, Atg7 and p62 levels (a-c), and measurement of LC3-immunopositive punctate signals and growth ability in soft agar (d and e). (f-h) Separately silencing Atg5, LC3B, Atg7, Atg4 or Atg16 each reversed the pro-autophagic and tumor-suppressive effects of AKT inhibition by an AKT inhibitor (AKT-in-IX) or a constitutively activated mutant (S253A) of FOXO3a (ca-FOXO3a) in CK1α-silenced PLLC-PTEN cells. (i-k) Separately silencing Atg5, LC3, Atg4 or Atg16 in Atg7-overexpressing NSCLC cells each abrogated Atg7-induced autophagy or suppression of tumor cell growth. (l-n) The effects of separately overexpressing Atg5, LC3, Atg7, Atg4 or Atg16 in CK1α-silenced PLLC-PTEN cells on autophagy or growth ability in soft agar were evaluated. All experiments were repeated three times with similar results and immunoblots in a-c, f, i and l are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01). Source data for precise P values are available in Supplementary Table 4.
Supplementary Figure 8 CK1α-induced aberrant autophagy in normal lung epithelial cells.
. (a) MTT assays revealing cell growth of NLE and BEAS-2B cells overexpressing wild-type CK1α or the ΔCK1α mutant. (b and c) The effect of silencing CK1α on autophagic flux in NLE and BEAS-2B cells in the absence or presence, respectively, of 10 nM Baf-A1 treatment. (d) WB analysis of changes in PTEN protein levels, and phosphorylation of AKT at ser473 and FOXO3a at ser253 in indicated cells. WB analysis of relevant proteins in indicated cells was performed. (e) WB analysis of changes in CK1α, LC3 conversion, PTEN, Atg7 and p62 levels, as well as FOXO3a phosphorylation at ser253 in indicated cells with indicated treatments. (f) Representative images of cell morphology of indicated cells. (g-i) NLEs or BEAS-2B cells were pre-silenced for CK1α or Atg7, followed by transduction of oncogenic HRasV12. Resultant cells were examined with cell survival/growth by MTT assay (g), with ultrastructural cytoplasimic autophagosomes and/or autolysosomes by TEM observation (h), and with autophagy by detecting LC3-immunopositive punctate signals (i). (j) Tumor tissues from xenografts of malignant cells transformed with cooperative activation of oncogenic HRasV12 and silencing CK1α or Atg7. (k) 3-D soft agar assays were performed to evaluate the malignantly transforming ability of indicated cells. Scale bar: 5 μm (c), 100 μm (f), 200 nm (h and j) and 5 μm (i). All experiments were repeated three times with similar results and images/immunoblots in b-f and h-k are representative of three independent experiments. Data represent mean +/- SD of n = 3 biologically independent experiments. A two-tailed Student’s t-test was used for statistical analysis (* P<0.05, ** P<0.01). Source data for precise P values are available in Supplementary Table 4.
Supplementary Figure 9
Unprocessed original scans of western blots presented in Figure 1b, 1d, 1h, 2b–k, 3b, 3i, 4a, 4b, 4f, 4g, 4i, 5a, 5k, 7d, 7e, 7g and 7i.
Supplementary information
Supplementary Information
Supplementary Figures 1–9 and legends for Supplementary Tables 1–4.
Rights and permissions
About this article
Cite this article
Cai, J., Li, R., Xu, X. et al. CK1α suppresses lung tumour growth by stabilizing PTEN and inducing autophagy. Nat Cell Biol 20, 465–478 (2018). https://doi.org/10.1038/s41556-018-0065-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0065-8
This article is cited by
-
RGS20 promotes non-small cell lung carcinoma proliferation via autophagy activation and inhibition of the PKA-Hippo signaling pathway
Cancer Cell International (2024)
-
FOXO3a-regulated arginine metabolic plasticity adaptively promotes esophageal cancer proliferation and metastasis
Oncogene (2024)
-
AKT2S128/CCTαS315/319/323-positive cancer-associated fibroblasts (CAFs) mediate focal adhesion kinase (FAK) inhibitors resistance via secreting phosphatidylcholines (PCs)
Signal Transduction and Targeted Therapy (2024)
-
Insights on E1-like enzyme ATG7: functional regulation and relationships with aging-related diseases
Communications Biology (2024)
-
Epigenetic and post-translational modifications in autophagy: biological functions and therapeutic targets
Signal Transduction and Targeted Therapy (2023)