Abstract
Changes in the micro-environment of fibrous connective tissue can lead to alterations in the phenotypes of tissue-resident cells, yet the underlying mechanisms are poorly understood. Here, by visualizing the dynamics of histone spatial reorganization in tenocytes and mesenchymal stromal cells from fibrous tissue of human donors via super-resolution microscopy, we show that physiological and pathological chemomechanical cues can directly regulate the spatial nanoscale organization and density of chromatin in these tissue-resident cell populations. Specifically, changes in substrate stiffness, altered oxygen tension and the presence of inflammatory signals drive chromatin relocalization and compaction into the nuclear boundary, mediated by the activity of the histone methyltransferase EZH2 and an intact cytoskeleton. In healthy cells, chemomechanically triggered changes in the spatial organization and density of chromatin are reversible and can be attenuated by dynamically stiffening the substrate. In diseased human cells, however, the link between mechanical or chemical inputs and chromatin remodelling is abrogated. Our findings suggest that aberrant chromatin organization in fibrous connective tissue may be a hallmark of disease progression that could be leveraged for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The authors declare that the main data supporting the findings in this study are available within the paper and its Supplementary Information. The raw STORM super-resolution data files are too large to be publicly shared but are available for research purposes from the corresponding authors on reasonable request. Source data are provided with this paper.
Code availability
The custom code for the super-resolution image analysis is available at https://github.com/melikelab/Su-Chin/tree/main/New%20folder.
References
Humphrey, J. D., Dufresne, E. R. & Schwartz, M. A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. Mol. Cell Biol. 15, 802–812 (2014).
Wang, N., Tytell, J. D. & Ingber, D. E. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 (2009).
Makris, E. A., Hadidi, P. & Athanasiou, K. A. The knee meniscus: structure–function, pathophysiology, current repair techniques, and prospects for regeneration. Biomaterials 32, 7411–7431 (2011).
Snedeker, J. G. & Foolen, J. Tendon injury and repair—a perspective on the basic mechanisms of tendon disease and future clinical therapy. Acta Biomater. 63, 18–36 (2017).
Han, W. M. et al. Microstructural heterogeneity directs micromechanics and mechanobiology in native and engineered fibrocartilage. Nat. Materials https://doi.org/10.1038/nmat4520 (2016).
Tsai, S. L., Nödl, M. T. & Galloway, J. L. Bringing tendon biology to heel: leveraging mechanisms of tendon development, healing, and regeneration to advance therapeutic strategies. Dev. Dyn. 250, 393–413 (2021).
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Ricci, M. A., Manzo, C., García-Parajo, M. F., Lakadamyali, M. & Cosma, M. P. Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158 (2015).
Walker, C. J. et al. Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-021-00709-w (2021).
Heo, S. J. et al. Mechanically induced chromatin condensation requires cellular contractility in mesenchymal stem cells. Biophys. J. 111, 864–874 (2016).
Heo, S. J. et al. Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. eLife 5, e18207 (2016).
Le, H. Q. et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18, 864–875 (2016).
Nava, M. M. et al. Heterochromatin-driven nuclear softening protects the genome against mechanical stress-induced damage. Cell 181, 800–817.e22 (2020).
Stephens, A. D. et al. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30, 2320–2330 (2019).
Lakadamyali, M. & Cosma, M. P. Visualizing the genome in high resolution challenges our textbook understanding. Nat. Methods https://doi.org/10.1038/s41592-020-0758-3 (20DOI20).
Ma, H. & Liu, Y. Super-resolution localization microscopy: toward high throughput, high quality, and low cost. APL Photonics 5, 060902 (2020).
Xu, J. et al. Super-resolution imaging reveals the evolution of higher-order chromatin folding in early carcinogenesis. Nat. Commun. https://doi.org/10.1038/s41467-020-15718-7 (2020).
Finnamore, E. et al. Transverse tendon stiffness is reduced in people with Achilles tendinopathy: a cross-sectional study. PLoS ONE https://doi.org/10.1371/journal.pone.0211863 (2019).
McBeath, R. et al. Tendinosis develops from age- and oxygen tension-dependent modulation of Rac1 activity. Aging Cell 18, e12934 (2019).
Shah, R. R., Nerurkar, N. L., Wang, C. C. & Galloway, J. L. Tensile properties of craniofacial tendons in the mature and aged zebrafish. J. Orthop. Res. 33, 867–873 (2015).
Cho, N. S., Hwang, J. H., Lee, Y. T. & Chae, S. W. Tendinosis-like histologic and molecular changes of the Achilles tendon to repetitive stress: a pilot study in rats. Clin. Orthop. Relat. Res. 469, 3172–3180 (2011).
Otterstrom, J. et al. Super-resolution microscopy reveals how histone tail acetylation affects DNA compaction within nucleosomes in vivo. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz593 (2019).
Heinz, K. S. et al. Peripheral re-localization of constitutive heterochromatin advances its replication timing and impairs maintenance of silencing marks. Nucleic Acids Res. 46, 6112–6128 (2018).
Shaklai, S., Amariglio, N., Rechavi, G. & Simon, A. J. Gene silencing at the nuclear periphery. FEBS J. 274, 1383–1392 (2007).
Heo, S. J. et al. Nuclear softening expedites interstitial cell migration in fibrous networks and dense connective tissues. Sci. Adv. https://doi.org/10.1126/sciadv.aax5083 (2020).
Kieffer-Kwon, K. R. et al. Myc regulates chromatin decompaction and nuclear Architecture during B cell activation. Mol. Cell https://doi.org/10.1016/j.molcel.2017.07.013 (2017).
Neguembor, M. V. et al. Transcription-mediated supercoiling regulates genome folding and loop formation. Mol. Cell 81, 3065–3081 (2021).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Killaars, A. R. et al. Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells. Adv. Sci. (Weinh) 6, 1801483 (2019).
LaCroix, A. S., Duenwald-Kuehl, S. E., Lakes, R. S. & Vanderby, R. Relationship between tendon stiffness and failure: a metaanalysis. J. Appl. Physiol. 115, 43–51 (2013).
Handorf, A. M., Zhou, Y., Halanski, M. A. & Li, W. J. Tissue stiffness dictates development, homeostasis, and disease progression. Organogenesis 11, 1–15 (2015).
Cosgrove, B. D. et al. N-cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat. Mater. 15, 1297–1306 (2016).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Bártová, E., Kreǰcí, J., Harničarová, A., Galiová, G. & Kozubek, S. Histone modifications and nuclear architecture: a review. J. Histochem. Cytochem. https://doi.org/10.1369/jhc.2008.951251 (2008).
Venkatesh, S. & Workman, J. L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015).
Liu, X. et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature https://doi.org/10.1038/nature19362 (2016).
Akkers, R. C. et al. A Hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in xenopus embryos. Dev. Cell 17, 425–434 (2009).
Gan, L. et al. Epigenetic regulation of cancer progression by EZH2: From biological insights to therapeutic potential. Biomark. Res. https://doi.org/10.1186/s40364-018-0122-2 (2018).
Dudakovic, A. et al. Epigenetic control of skeletal development by the histone methyltransferase Ezh2. J. Biol. Chem. https://doi.org/10.1074/jbc.M115.672345 (2015).
Heo, S.-J. et al. Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Sci. Rep. 5, 16895 (2015).
Sato, T. et al. Transcriptional selectivity of epigenetic therapy in cancer. Cancer Res. https://doi.org/10.1158/0008-5472.CAN-16-0834 (2017).
Liu, Y.-B. et al. LPA induces osteoblast differentiation through interplay of two receptors: LPA1 and LPA4. J. Cell. Biochem. https://doi.org/10.1002/jcb.22471 (2010).
Toews, M. L., Ustinova, E. E. & Schultz, H. D. Lysophosphatidic acid enhances contractility of isolated airway smooth muscle. J. Appl. Physiol. 83, 1216–1222 (1997).
Voorhees, P. W. The theory of Ostwald ripening. J. Stat. Phys. 38, 231–252 (1985).
Guvendiren, M. & Burdick, J. A. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. https://doi.org/10.1038/ncomms1792 (2012).
Caliari, S. R. et al. Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci. Rep. 6, 21387 (2016).
Dakin, S. G. et al. Inflammation activation and resolution in human tendon disease. Sci. Transl. Med. 7, 311ra173 (2015).
Tempfer, H. & Traweger, A. Tendon vasculature in health and disease. Front. Physiol. 6, 330 (2015).
Millar, N. L. et al. Hypoxia: a critical regulator of early human tendinopathy. Ann. Rheum. Dis. 71, 302–310 (2012).
John, T. et al. Effect of pro-inflammatory and immunoregulatory cytokines on human tenocytes. J. Orthop. Res. 28, 1071–1077 (2010).
Mobasheri, A. & Shakibaei, M. Is tendinitis an inflammatory disease initiated and driven by pro-inflammatory cytokines such as interleukin 1β? Histol. Histopathol. 28, 955–964 (2013).
Svensson, R. B., Heinemeier, K. M., Couppé, C., Kjaer, M. & Magnusson, S. P. Effect of aging and exercise on the tendon. J. App. Physiol. 121, 1237–1246 (2016).
Turan, A., Teber, M. A., Yakut, Z. I., Unlu, H. A. & Hekimoglu, B. Sonoelastographic assessment of the age-related changes of the Achilles tendon. Med. Ultrasonography 17, 58–61 (2015).
Arya, S. & Kulig, K. Tendinopathy alters mechanical and material properties of the Achilles tendon. J. Appl. Physiol. 108, 670–675 (2010).
Arvind, V. & Huang, A. H. Reparative and maladaptive inflammation in tendon healing. Front. Bioeng. Biotechnol. 9, 719047 (2021).
Schulze-Tanzil, G. et al. The role of pro-inflammatory and immunoregulatory cytokines in tendon healing and rupture: new insights. Scand. J. Med. Sci. Sports 21, 337–351 (2011).
Mirabella, A. C., Foster, B. M. & Bartke, T. Chromatin deregulation in disease. Chromosoma 125, 75–93 (2016).
Zoghbi, H. Y. & Beaudet, A. L. Epigenetics and human disease. Cold Spring Harb. Persp. Biol. 8, a019497 (2016).
Saul, D. & Kosinsky, R. L. Epigenetics of aging and aging-associated diseases. Int. J. Mol. Sci. 22, 401 (2021).
Gnan, S., Liu, Y., Spagnuolo, M. & Chen, C.-L. The impact of transcription-mediated replication stress on genome instability and human disease. Genome Instab. Dis. 1, 207–234 (2020).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Becker, J. S. et al. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol. Cell https://doi.org/10.1016/j.molcel.2017.11.030 (2017).
van Steensel, B. & Belmont, A. S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017).
Bitman-Lotan, E. & Orian, A. Nuclear organization and regulation of the differentiated state. Cell. Mol. Life Sci. https://doi.org/10.1007/s00018-020-03731-4 (2021).
Yao, L., Bestwick, C. S., Bestwick, L. A., Maffulli, N. & Aspden, R. M. Phenotypic drift in human tenocyte culture. Tissue Eng. 12, 1843–1849 (2006).
Gardner, O. F. W., Alini, M. & Stoddart, M. J. Mesenchymal stem cells derived from human bone marrow. Methods Molec. Biol. 1340, 41–52 (2015).
Loebel, C., Mauck, R. L. & Burdick, J. A. Local nascent protein deposition and remodelling guide mesenchymal stromal cell mechanosensing and fate in three-dimensional hydrogels. Nat. Mater. https://doi.org/10.1038/s41563-019-0307-6 (2019).
Song, K. H. et al. Influence of fiber stiffness on meniscal cell migration into dense fibrous networks. Adv. Healthc. Mater. https://doi.org/10.1002/adhm.201901228 (2020).
Davidson, M. D. et al. Engineered fibrous networks to investigate the influence of fiber mechanics on myofibroblast differentiation. ACS Biomater. Sci. Eng. 5, 3899–3908 (2019).
Sagui, C. & Desai, R. C. Ostwald ripening in systems with competing interactions. Phys. Rev. Lett. 74, 1119–1122 (1995).
Cahn, J. W. & Hilliard, J. E. Free energy of a nonuniform system. I. Interfacial free energy. The J. Chem. Phys. 28, 258 (1958).
Zwicker, D., Hyman, A. A. & Jülicher, F. Suppression of Ostwald ripening in active emulsions. Phys. Rev. E 92, 012317 (2015).
Poleshko, A. et al. Genome-nuclear lamina interactions regulate cardiac stem cell lineage restriction. Cell 171, 573–587 (2017).
Buchwalter, A., Kaneshiro, J. M. & Hetzer, M. W. Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet. 20, 39–50 (2019).
Towbin, B. D., Gonzalez-Sandoval, A. & Gasser, S. M. Mechanisms of heterochromatin subnuclear localization. Trends Biochem. Sci. 38, 356–363 (2013).
Glotzer, S. C., Stauffer, D. & Jan, N. Glotzer, Stauffer, and Jan reply. Phys. Rev. Lett. 75, 1675 (1995).
Christensen, J. J., Elder, K. & Fogedby, H. C. Phase segregation dynamics of a chemically reactive binary mixture. Phys. Rev. E 54, R2212–R2215 (1996).
Alisafaei, F., Jokhun, D. S., Shivashankar, G. V. & Shenoy, V. B. Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc. Natl Acad. Sci. USA 116, 13200–13209 (2019).
Cella Zanacchi, F. et al. A DNA origami platform for quantifying protein copy number in super-resolution. Nat. Methods 14, 789–792 (2017).
Andronov, L., Orlov, I., Lutz, Y., Vonesch, J. L. & Klaholz, B. P. ClusterViSu, a method for clustering of protein complexes by Voronoi tessellation in super-resolution microscopy. Sci. Rep. 6, 24084 (2016).
Levet, F. et al. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods https://doi.org/10.1038/nmeth.3579 (2015).
Acknowledgements
The authors thank K.H. Song for assistance with the design of the microfluidic chamber, E. Sorokina for assistance with the preparation of reagents for STORM imaging and P. K. Relich for help with data visualization and analysis tools. The research was funded by the National Institutes of Health (grant no. K01 AR07787 to S-J.H.), Penn Center for Musculoskeletal Disorders (grant no. P30 AR069619 to S-J.H, M.L. and R.L.M.), Department of Veterans Affairs (grant no. IK6 RX003416 to R.L.M.), R01 GM133842 (to M.L.), U01DA052715 (to M.L.) and NSF Science and Technology Center for Engineering Mechanobiology (grant no. CMMI-1548571 to M.L., R.L.M and V.S.). The theoretical work was supported by National Cancer Institute Awards R01CA232256 and U54CA261694; National Institute of Biomedical Imaging and Bioengineering Awards R01EB017753 and R01EB030876; and NSF Grants MRSEC/DMR-1720530 and DMS-1953572.
Author information
Authors and Affiliations
Contributions
S.-J.H., S.T., X.C., C.L., B.X., R.M., J.A.B., V.B.S., R.L.M. and M.L. designed the studies, and analysed and interpreted the data. S.-J.H., S.T., X.C., C.L. and B.X. performed the experiments. S.-J.H., R.L.M. and M.L. drafted the manuscript, and all authors edited the final submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Guanbin Song and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
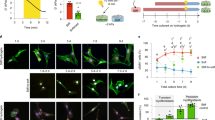
Extended Data Fig. 1 Custom-PDMS microfluidic chamber, and changes in chromatin distribution.
(a) Picture and schematic of a custom-PDMS microfluidic fluid shear stress (FSS) device. (b) Representative STORM super-resolution images of H2B rendered as a density map showing redistribution of H2B in hMSCs in response to FSS of varying magnitude 1 ~ 5 dyne/cm2) and duration (0.5 ~ 2 h). (c) Quantification of changes in the ratio of the total number of H2B localizations per unit area at the nuclear border to the total number of H2B localizations at the inner part of the nucleus with/without the application of FSS (*: p < 0.05 vs. Ctrl, +: p < 0.05 vs. 1D/0.5 h, a: p < 0.05 vs. 5D/0.5 h). The values have been normalized to Ctrl cells. (d) Changes in chromatin condensation with the application of FSS in (d) sparse chromatin compartment or (e) dense chromatin compartment in hMSCs. The Voronoi polygon density of the dense or the sparse chromatin compartment in cells subjected to FSS is shown normalized to the Voronoi polygon density in the absence of FSS (n = 5 nuclei/group, *: p < 0.001 vs. Ctrl, +: p < 0.001 vs. 1D/0.5 h, a: p < 0.001 vs. 5D/0.5 h, one-way ANOVA). Experiments were carried out at least in duplicate. Error bars, means ± s.d.
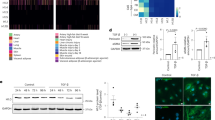
Extended Data Fig. 2 Changes in chromatin condensation with substrate stiffness in sparse chromatin or dense chromatin compartments.
a,b, The Voronoi polygon density of the sparse (a) or dense (b) chromatin compartment is shown normalized to the Voronoi polygon density in human tenocytes grown on Glass, showing a decrease/increase in the condensation of dense chromatin compartment on stiff/soft substrates (n = 5 nuclei/group, *: p < 0.001 vs. glass, +: p < 0.001 vs. soft, one-way ANOVA). Changes in chromatin condensation in sparse chromatin (c) or dense chromatin (d) compartments. The Voronoi polygon density of the sparse or dense chromatin compartment is shown normalized to the Voronoi polygon density in human young tenocytes grown under normoxic (N) conditions, showing an increase in the condensation of dense chromatin under hypoxic (H) conditions (n = 5 nuclei/group, *: p < 0.001 vs. young healthy tenocyte under the normoxic conditions, +: p < 0.001 vs. tendinosis tenocyte under the normoxic conditions, one-way ANOVA). Changes in chromatin condensation with exposure to inflammatory cytokines in sparse chromatin (e) or dense chromatin (f) compartments. The Voronoi polygon density of the sparse or dense chromatin compartment is shown normalized to the Voronoi polygon density in human young tenocytes without treatment with inflammatory cytokines, showing an increase in the condensation of dense chromatin upon treatment (n = 5 nuclei/group, *: p < 0.001 vs. young human tenocyte control, +: p < 0.001 vs. tendinosis young human tenocyte, One-way ANOVA). Error bars, means ± s.d.
Supplementary information
Supplementary Information
Supplementary methods, figures and tables.
Supplementary Data 1
Source data and statistics for Supplementary Fig. 3.
Supplementary Data 2
Source data and statistics for Supplementary Fig. 6b.
Supplementary Data 3
Source data and statistics for Supplementary Fig. 15a.
Source data
Source Data Fig. 1
Source data and statistics.
Source Data Fig. 2
Source data and statistics.
Source Data Fig. 3
Source data and statistics.
Source Data Fig. 4
Source data and statistics.
Source Data Fig. 5
Source data and statistics.
Source Data Fig. 6
Source data and statistics.
Source Data Fig. 7
Source data and statistics.
Source Data Extended Data Fig. 1
Source data and statistics.
Rights and permissions
About this article
Cite this article
Heo, SJ., Thakur, S., Chen, X. et al. Aberrant chromatin reorganization in cells from diseased fibrous connective tissue in response to altered chemomechanical cues. Nat. Biomed. Eng 7, 177–191 (2023). https://doi.org/10.1038/s41551-022-00910-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-022-00910-5
This article is cited by
-
Fluorescence-based super-resolution-microscopy strategies for chromatin studies
Chromosoma (2023)
-
InterLINCing Chromatin Organization and Mechanobiology in Laminopathies
Current Cardiology Reports (2023)
-
Chromatin reprogramming and bone regeneration in vitro and in vivo via the microtopography-induced constriction of cell nuclei
Nature Biomedical Engineering (2023)