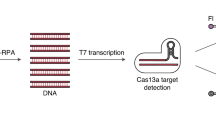
Abstract
CRISPR-based assays for the detection of nucleic acids are highly specific, yet they are not fast, sensitive or easy to use. Here we report a one-step fluorescence assay for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in nasopharyngeal samples, with a sample-to-answer time of less than 20 minutes and a sensitivity comparable to that of quantitative real-time PCR with reverse transcription (RT–qPCR). The assay uses suboptimal protospacer adjacent motifs, allowing for flexibility in the design of CRISPR RNAs and slowing down the kinetics of Cas12a-mediated collateral cleavage of fluorescent DNA reporters and cis cleavage of substrates, which leads to stronger fluorescence owing to the accumulation of amplicons generated by isothermal recombinase polymerase amplification. In a set of 204 nasopharyngeal samples with RT–qPCR cycle thresholds ranging from 18.1 to 35.8, the assay detected SARS-CoV-2 with a sensitivity of 94.2% and a specificity of 100%, without the need for RNA extraction. Rapid and sensitive assays for nucleic acid testing in one pot that allow for flexibility in assay design may aid the development of reliable point-of-care nucleic acid testing.
Similar content being viewed by others
Main
In recent decades, there have been many large-scale outbreaks of epidemic diseases caused by viruses such as severe acute respiratory syndrome coronavirus (SARS-CoV)1,2, Middle East respiratory syndrome coronavirus (MERS-CoV)3, human immunodeficiency virus (HIV)4,5, Zika virus6 and Ebola virus7, as well as the current SARS-CoV-2 (coronavirus disease 2019 (COVID-19)) pandemic8,9. As of now, the world is facing a serious challenge in controlling the spread of SARS-CoV-2, which has caused several million deaths and tremendous economic losses8,9. The spread of SARS-CoV-2 has been rapid, in part due to the high prevalence of presymptomatic and asymptomatic transmission10,11,12. Exploring rapid, specific and sensitive diagnostic strategies is essential for combating the COVID-19 pandemic.
However, the availability of nucleic acid diagnostic tests has been unable to keep pace with the accelerated spread, as quantitative real-time PCR with reverse transcription (RT–qPCR)-based tests, the gold standard for SARS-CoV-2 diagnosis, require skilled personnel and equipment infrastructure, and also have long sample-to-answer times13,14,15,16. Point-of-care nucleic acid tests that are sensitive enough to detect the virus in asymptomatic carriers and have turnaround times that are fast enough are critical to control the pandemic. Isothermal amplification assays, such as recombinase polymerase amplification (RPA)17,18 and loop-mediated isothermal amplification (LAMP)19,20, provide rapid and low-cost alternatives. Abbott’s ID NOW COVID-19 test, which utilizes isothermal amplification for unextracted samples, is able to report results in 13 min; however, this test showed negative results for a third to 45% of the samples that tested positive by RT–qPCR21. Moreover, isothermal amplification assays are often subject to nonspecific amplification, resulting in false-positive results22,23,24,25.
CRISPR–Cas (clustered regularly interspaced short palindromic repeats–CRISPR-associated proteins) are endonucleases guided by RNA to cleave specific nucleic acid sequences, the process being known as cis cleavage26,27,28,29. Upon recognizing sequences, Cas12 and Cas13 exhibit collateral activity to cleave non-target single-stranded (ss) DNA and ssRNA, respectively, the process being known as trans cleavage26,27,28,29. Amplification of target sequences and sequential cis cleavage by Cas12 or Cas13 family member proteins, followed by trans cleavage of caged reporter molecules created Cas13a or Cas13b-based SHERLOCK (Specific High Sensitivity Enzymatic Reporter Unlocking) and Cas12a-based DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter)26,27,28,30,31. Both SHERLOCK and DETECTR have been clinically validated to detect SARS-CoV-232,33. However, these approaches use a two-step process (amplification of target sequences followed by CRISPR detection), in addition to an RNA extraction step32,33. The multiple liquid-handling steps increase time of operation and risk of cross-contamination32,33. SHERLOCK has been further developed as STOPCovid.v1 (SHERLOCK Testing in One Pot, version 1) and SHINE (SHERLOCK and HUDSON Integration to Navigate Epidemics) to address this issue; in these assays, combined isothermal amplification and CRISPR detection were used to test unextracted samples in one step22,23. The simplicity of STOPCovid.v1 and SHINE makes them attractive for point-of-care testing22,23. However, they are much less sensitive than RT–qPCR, and require a total reaction time of approximately 1 h22,23. A method for amplification-free detection of SARS-CoV-2 using Cas13a was reported to get test results in 30 min, but it is at least 100-fold less sensitive than RT–qPCR34.
Here we develop a method based on Cas12a, which we named sPAMC (for suboptimal PAM (protospacer adjacent motifs) of Cas12a-based test with enhanced flexibility, speed, sensitivity and reproducibility). We found that in one-step CRISPR detection where isothermal amplification and cleavage of Cas12a occur simultaneously in the same tube, the use of CRISPR RNAs (crRNAs) targeting substrates with suboptimal PAMs rather than the routinely used canonical PAMs could accelerate the reaction speed by 2- to 3-fold. Moreover, extensive testing showed that sPAMC exhibited greater sensitivity and reliability than the one-pot test using canonical PAMs. The high abundance of suboptimal PAM sequences makes this method more flexible for optimization. sPAMC allows for the detection of a human cytomegalovirus (HCMV) DNA virus in 10 min and of the SARS-CoV-2 RNA virus in 15 min using unextracted samples, with comparable detection limit as qPCR in both cases. sPAMC is a CRISPR-mediated detection method that exhibits the combined advantages of being simple, fast, sensitive, reliable and flexible. It is easy to use (unextracted samples in a one-pot reaction) and fast (showing comparable speed as isothermal amplification alone). Furthermore, it is as sensitive, reliable and flexible as RT–qPCR. We also provide a mechanistic understanding of how to improve the one-pot reaction, providing a path to continuously improve the performance of CRISPR detections using Cas12a and other effectors, and to apply CRISPR detection beyond lab use to point-of-care diagnostics.
Results
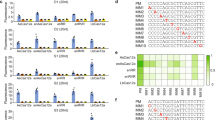
We designed a number of crRNAs targeting the replicase polyprotein 1ab (Orf1ab) and envelope (E) genes of SARS-CoV-2 for a one-pot reaction where RPA-based amplification and Cas12a-mediated detection occurred simultaneously. We noticed that several crRNAs displayed faster kinetics of fluorescence signals than others in the one-pot reaction (Extended Data Fig. 1a,b). Detailed analysis revealed that those crRNAs with better performance are all designed to use a suboptimal PAM of Cas12a (NTTV and TTNT) rather than the canonical PAM (TTTV) (Extended Data Fig. 1c). To determine whether suboptimal PAM is the key to enhance Cas12a-mediated detection in a one-pot reaction, the substrates of spacers 4 and 5 targeting the Orf1ab gene (named as spacers 4 and 5 hereafter) were mutated from suboptimal PAM to the canonical PAM. As expected, crRNAs using suboptimal PAM generated weaker and slower collateral activity than those with the canonical PAM (Fig. 1a,b). We then compared these substrates in one-pot reactions. In contrast to the collateral activity assay, in the one-pot reaction, both spacers 4 and 5 using suboptimal PAM exhibited faster kinetics than the canonical PAM (Fig. 1c,d).
a–d, The fluorescence signal of Orf1ab gene spacer 4 and spacer 5 in collateral activity tests (a,b) and one-pot reactions (c,d) at 37 °C. Suboptimal PAMs for Orf1ab spacer 4 (GTTG) and spacer 5 (CTTA) were mutated to canonical PAMs for spacer 4 (TTTG) and spacer 5 (TTTA), respectively. e–h, Summary map of fluorescent kinetics for positions 1–3 point-mutated suboptimal PAMs and three canonical PAMs in collateral activity test (e,f) and the corresponding one-pot reaction of spacer 4 (g) and spacer 5 (h). Time to half-maximum fluorescence was determined. Fluorescence values were determined at 40 and 20 min for collateral activities and one-pot reactions, respectively. The concentrations of dsDNA substrates were 3.5 nM in collateral activity tests and 2,340 fM in one-pot reactions. a,c,e,g, Orf1ab spacer 4. b,d,f,h, Orf1ab spacer 5. Mean ± s.d. of n = 3 technical replicates for e–h. NC represents reactions without substrate in a–d.
To explore which types of suboptimal PAMs exhibited faster reactions in the one-pot reactions, substrates of spacers 4 and 5 of the Orf1ab gene, spacer 2 of Spike (S) gene of SARS-CoV-2 and spacer 1 of the human papillomavirus type 18 (HPV18) L1 gene were point-mutated from TTTV to VTTV, TVTV or TTVV. A comparison of the collateral activity and one-pot reaction for 120 suboptimal PAMs of 4 spacers indicated that more than 80% of spacers with suboptimal PAMs showed a faster reaction than those with the canonical PAM in the one-pot reaction, and most of the suboptimal PAMs with better performance were VTTV, TCTV and TTVV (Fig. 1e–h, Extended Data Fig. 2a–d and Supplementary Figs. 1–4). The protein structure of Cas12a shows that the PAM-interacting domain mainly contacts the second nucleotide of the target strand; therefore, mutating the second nucleotide of PAM from pyrimidine to purine is likely to dramatically impair the activity of Cas12a (Supplementary Fig. 5). Indeed, some TATV and TGTV PAMs, but not TCTV PAMs, showed slower kinetics with reduced fluorescence signals in the one-pot reaction; and consistently, these suboptimal PAMs all demonstrated much lower collateral activity than the canonical PAMs (Supplementary Figs. 1–4). For spacers 4 and 5, TTTT PAM exhibited faster kinetics than TTTV PAM in the one-pot reaction, indicating that the fourth nucleotide of the PAM may also be modified to tune the activity of Cas12a (Supplementary Fig. 6a–d). We then introduced two point mutations into the PAM. The introduction of two PAM point mutations (TTTV to TTVT) for spacer 4 and part of spacer 5, produced faster kinetics in the one-pot reaction (Supplementary Fig. 6a–d). We then examined other suboptimal PAMs bearing two point mutations. The VVTV and VTVV PAMs for spacers 4 and 5 showed reduced kinetics and signals in the one-pot reaction and collateral activity (Supplementary Fig. 6e–h). Interestingly, mutation of TTTV to TCCV produced a superior reaction in the one-pot reaction (Supplementary Fig. 6h). This may be because the PAM of Cas12a can tolerate the T to C mutation; TCCV has been characterized as a functional suboptimal PAM35. CCCV, with three point mutations, was also recognized as a suboptimal PAM35 and indeed, CCCA was faster than the canonical PAM in the one-pot reaction (Supplementary Fig. 6i,j). As a negative control, another sequence with three point mutations of the PAM sequences, AGCA, showed minimal activity in the one-pot reaction. Together, these results suggest that a delicate level of collateral activity is associated with the performance of one-pot reaction. Here we conclude that most VTTV, TCTV and TTVV, as well as some TRTV, TTNT and YYYN (except TTTV) PAMs, outperform the canonical PAM in the one-pot reaction.
To further demonstrate that the use of suboptimal PAM sequences could accelerate the one-pot reaction, we synthesized crRNAs targeting the E and S genes of SARS-CoV-2. These crRNAs demonstrated faster reactions on substrates with suboptimal PAMs than on those with canonical PAMs in the one-pot reaction, indicating that the use of suboptimal PAM could be a general strategy to accelerate the speed of the Cas12a-based one-pot test (Extended Data Fig. 3a,b).
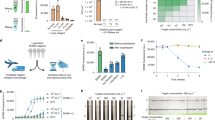
Previous studies indicate that although one-pot CRISPR diagnostics are simple to operate, their sensitivity is lower than that of two-step methods, in which target amplification and CRISPR detection are performed sequentially33. We therefore investigated whether the application of suboptimal PAM could improve the sensitivity of the one-pot test. The detection limit of spacer 4 using canonical PAM was 234 fM concentration of double-stranded DNA (dsDNA) in the one-pot reaction; in contrast, its detection limit using suboptimal PAM was 2.34 fM concentration of dsDNA (Fig. 2a,b). To compare the reliability of tests using suboptimal and canonical PAMs, we repeated the experiments under identical conditions 10 times with 2 replicates each time. With substrates (2,340 fM concentration of dsDNA) and incubation times sufficient for both suboptimal and canonical PAMs, the fluorescence signal from the suboptimal PAM group was highly consistent across all replicates; in contrast, signals from the canonical PAM group varied more than 10-fold across replicates (Fig. 2c). We then compared the detection limit and reliability of an additional 2 crRNAs using canonical or suboptimal PAMs. Both crRNAs using suboptimal PAMs exhibited a ~10- to 100-fold increase in sensitivity and much more consistent signal production compared with those with canonical PAMs, demonstrating the improved sensitivity and reliability of suboptimal PAM in the one-pot reaction (Fig. 2d–i).
The sensitivity and reliability of one-pot reactions using suboptimal PAMs and canonical PAMs were compared. crRNAs targeting the Orf1ab gene (spacers 4 and 5) and envelope (E) gene (spacer 8) of SARS-CoV-2 were used. a,b, The sensitivity of spacer 4 using suboptimal PAM (a) and canonical PAM (b). c, The reliability of spacer 4 using each PAM. d,e, The sensitivity of spacer 5 using suboptimal PAM (d) and canonical PAM (e). f, The reliability of spacer 5 using each PAM. g,h The sensitivity of spacer 8 using suboptimal PAM (g) and canonical PAM (h). i, The reliability of spacer 8 using each PAM. For c, f and i, substrate concentrations were 2,340 fM, 2,340 fM and 325.5 fM, respectively; fluorescence values were determined 50 min after incubation, and the data are from 10 experiments with 2 replicates per experiment. For a, b, d, e, g and h, each experiment was repeated three times, with one representative result shown. For a–i, the reaction temperature was 37 °C.
To investigate the dose effect of Cas12a/crRNA ribonucleoprotein (RNP) in the one-pot reaction, we tested RNP doses ranging from 5.5 nM to 132 nM with assays using suboptimal or canonical PAMs. Reactions with suboptimal PAM showed stable kinetic curves and consistent results with RNP dose ranging from 22–132 nM (Supplementary Fig. 7a), whereas reactions with canonical PAM displayed drastic fluctuations in kinetic curves and highly variable signals, with even a minor change in RNP dose (Supplementary Fig. 7b). These data further elucidate that using a suboptimal PAM is key to reproducible results in the Cas12a-mediated one-pot test.
We next sought to understand the mechanism underlying the robust performance of suboptimal PAM-mediated one-pot detection. In the one-pot reaction, CRISPR detection and isothermal amplification might compete against each other, and the ultimate detection signal could rely on target amplification to generate adequate substrates for CRISPR detection. It is possible that crRNA using suboptimal PAM has slower initial kinetics of CRISPR detection and thus biases the reaction towards isothermal amplification. To examine this possibility, we monitored amplicon generation in the one-pot reaction. For spacer 4, the target amplicon was first observed 2 min after a one-pot reaction using suboptimal PAM, whereas in the canonical PAM group, it took 8–10 min to identify the amplicon (Fig. 3a). In addition, the amounts of amplicons generated by one-pot reactions with suboptimal PAM or RPA alone were much greater than those generated by one-pot reactions with canonical PAM at each time point (Fig. 3a). Consistently, the generation of amplicons by spacer 5 and two additional spacers also displayed faster kinetics and higher amounts of amplicons in tests with suboptimal PAM than those with canonical PAM, indicating a stronger interference with the RPA amplification when using canonical PAM (Fig. 3b and Supplementary Fig. 8a,b). To further compare the ability of isothermal amplification under the surveillance of Cas12a, one-pot reactions were carried out using mixed substrates composed of 50% suboptimal and 50% canonical PAMs. Amplicon sequencing analysis revealed that within the first minute of reaction, the amplicons from the suboptimal PAM substrate accounted for more than ~90% of the amplification products, supporting the perspective that the use of a suboptimal PAM is crucial in promoting RPA amplification under the pressure of competing Cas12a cleavage (Fig. 3c,d).
a,b, The accumulation of RPA amplicons in one-pot reactions. Components of RPA at concentrations of 33 nM crRNA/Cas12a RNP and 2,340 fM dsDNA substrates were incubated at 37 °C for 0, 2, 4, 6, 8, 10, 12, 14, 16, 18 or 20 min, and the resulting RPA amplicons were analysed in agarose gels. Arrows indicate amplicon products. c,d, Amplification and consumption of amplicons in one-pot reactions. Concentrations of 1,170 fM substrates containing suboptimal or canonical PAM were mixed at a ratio of 1:1 in one-pot reactions at 37 °C, and the percentage of each sequence at 0, 1, 3, 5, 7, 10, 15 and 20 min was determined by deep sequencing. n = 5 for each time point. e,f, In vitro cleavage activities of spacers 4 targeting substrates with suboptimal (GTTG) or canonical (TTTG) PAMs and spacers 5 targeting substrates with suboptimal (CTTA) or canonical (TTTA) PAMs. The crRNA/LbCas12a complex was incubated with dsDNA substrate at 37 °C for 0, 0.5, 1, 2, 5, 10, 15 or 20 min to determine the cis-cleavage activity. S, substrate; P, product. g,h, The binding affinity of RNP for dsDNA containing suboptimal or canonical PAMs was determined. crRNA/deactivated LbCas12a (dCas12a) complexes (0, 12.5, 25, 50, 100, 200 and 400 nM) were incubated with 5 nM dsDNA at 37 °C for 20 min, and EMSAs were performed to determine the bound and unbound portions. a,c,e,g, Orf1ab spacer 4. b,d,f,h, Orf1ab spacer 5. Each experiment was repeated three times, and data for one representative are shown.
Cas12a-mediated substrate binding and subsequent cis cleavage may interfere with RPA amplification. A time course of cis-cleavage activity for a constant amount of DNA substrates showed that cleavage of the canonical PAM substrates was completed within 30 s, whereas it took 10–20 min to complete cleavage of the suboptimal PAM substrates (Fig. 3e,f). Cas12a was able to bind suboptimal PAM substrates with reduced affinity35. We reasoned that delayed cleavage was due to weak binding of Cas12a to the DNA substrate with suboptimal PAM. In agreement, the electrophoretic mobility shift assay (EMSA) analysis of Cas12a binding affinity showed reduced binding with suboptimal PAM substrates compared with canonical PAM for both spacers 4 and 5 (Fig. 3g,h).
To further clarify the mechanism involved, we evaluated cis-cleavage activities, collateral activities and one-pot reaction of 120 PAMs (including suboptimal PAMs and canonical PAMs) for 4 different spacers (HPV18 L1 gene spacer 1, Orf1ab spacer 4, Orf1ab spacer 5 and S gene spacer 2) (Supplementary Figs. 1–4 and 9). We identified that Kcleave and the performance of one-pot reactions had clear correlation (Fig. 4a and Supplementary Table 1). All 12 canonical PAMs of these 4 spacers and 1 suboptimal PAM of Orf1ab spacer 4 had high Kcleave of 1.2–3.5 min−1. They performed well in collateral activity, but all had poor performance in one-pot reactions (Fig. 4a and Supplementary Table 1). The suboptimal PAMs with minimal cis cleavage (Kcleave = 0–0.1 min−1) had the worst performance in both collateral activity and one-pot reaction. In contrast, the suboptimal PAMs with intermediate Kcleave of 0.1–1.2 min−1 outperformed their respective canonical PAMs in one-pot reactions (Fig. 4a and Supplementary Table 1). These results indicate that the efficiency of cis cleavage is the key factor determining the performance of one-pot reactions. Due to excessive substrate consumption caused by canonical PAMs-mediated cis cleavage, the amplicon accumulation is slow and unstable, resulting in delayed or lack of collateral activity in the one-pot reaction. Although suboptimal PAMs with minimal cis cleavage allow accumulation of amplicon (substrates), they cannot execute sufficient collateral activity. In contrast, suboptimal PAMs with intermediate Kcleave allow substrate accumulation at the early stage of isothermal reaction while maintaining considerable collateral activity. We ranked the best performing suboptimal PAMs on the basis of the time to half-maximum fluorescence in one-pot reactions (Supplementary Table 1). In the top 5 best performing suboptimal PAMs for each spacer (20 PAMs in total), there are 12 VTTV and 5 TCTV. Therefore, we suggest that VTTV can be selected as the top suboptimal PAMs, and TCTV are good candidates for one-pot reactions.
a, Correlation of one-pot reaction and cis cleavage of 120 PAMs. Black dots represent canonical PAMs. Red dots represent suboptimal PAMs with better performance and blue dots represent suboptimal PAMs with comparable or worse performance than canonical PAMs in one-pot reactions. The unit of one-pot reaction (x axis) is defined as the time to half-maximum fluorescence (min) × an adjusted ratio based on the plateau signal of each PAM. This ratio is the value of the highest plateau fluorescence among 120 PAMs divided by the plateau fluorescence value of each PAM. The 3 suboptimal PAMs out of the 30 min range in the x axis still outperformed their corresponding canonical PAMs. One suboptimal PAM had similar Kcleave value as canonical PAMs, and it performed similarly as canonical PAMs in the one-pot reaction. b, Schematic workflow of amplification and cleavage in one-pot reactions. For substrates containing canonical PAM, cleavage is predominant in the initial stage of the reaction, resulting in excessive consumption of the dsDNA activator. In contrast, as amplification outcompetes cleavage for substrates containing suboptimal PAM, amplicons accumulate to stimulate faster and stronger fluorescence signal production. The up arrows represent the dominant process in the one-pot reactions.
Taken together, these data suggest a model for how suboptimal PAM functions to promote isothermal amplification and thus results in reliable and sensitive detection in a one-pot reaction (Fig. 4b). Given that CRISPR detection and isothermal amplification of substrates compete against each other, the decreased binding affinity of Cas12a for suboptimal PAM substrates promotes a shift of the balance from cleavage towards amplification and thus generates sufficient amplicons for detection; in contrast, the stronger binding affinity of canonical PAM allows cleavage to outcompete amplification and leads to delay or lack of amplicon production, which is responsible for the observed delay and instability of detection, with reduced sensitivity.
We have demonstrated that using suboptimal PAM is superior for one-pot reactions. These results were obtained using an RPA kit (AMP future). To examine whether this conclusion is valid using a different RPA kit, we performed experiments with an RPA kit from TwistDx. These two RPA kits showed no difference in amplification sensitivity (Supplementary Fig. 10a). However, when we combined RPA and Cas12a in one reaction, the TwistDx kit showed much less sensitivity and reduced fluorescence signal (Supplementary Fig. 10b). Results suggest that Cas12a is not fully compatible with the buffer environment of TwistDx RPA. A recent study showed that LwaCas13a was also not fully compatible with the TwistDx RPA buffer environment23. A method named SHINE was developed to retain both RPA and Cas13a activities in optimized buffers23. Inspired by this study, we increased RNP doses from 33.3 nM to 100 nM, 200 nM and 333 nM in the reaction. We found that 3–6-fold more RNP can substantially improve the fluorescence curve of one-pot reactions (Supplementary Fig. 10c). Using this improved condition and TwistDx kit, we compared the performance of canonical PAMs and suboptimal PAMs of 4 spacers. Similar to the AMP future kit, the suboptimal PAMs performed much better than canonical PAMs when both were using TwistDx kit (Supplementary Fig. 10d–g).
We have thus developed the sPAMC with enhanced flexibility, speed, sensitivity and reproducibility. To determine whether sPAMC is able to detect DNA viruses, we tested the method with human cytomegalovirus (HCMV), a double-stranded DNA virus. We first used a plasmid containing the UL55 gene sequence of HCMV as a substrate to compare the sensitivity of sPAMC with qPCR. Both assays showed the same detection limit of 5.953 × 10−4 amol plasmid per reaction (equal to 29.765 aM concentration in qPCR and 19.843 aM in sPAMC) (Fig. 5a,b). The fluorescence signal of sPAMC started to appear at approximately 6–10 min and reached a half-maximum at approximately 9–15 min for all concentrations tested (Fig. 5b). Notably, this speed is at least 2- to 3-fold faster than that of all published CRISPR-mediated one-pot tests with target amplification22,23,33,36. We then used sPAMC to measure the HCMV viral samples. The results showed a detection limit of 24 copies per reaction, comparable to that of qPCR (Fig. 5c,d). To enable a broader application of sPAMC, we used simple UV light instead of fluorescence spectroscopy to measure the signal. At the 10 min time point, all except the lowest viral concentration showed positive signals on UV detection, and at 15 min, the sample with the lowest number of viral copies (equal to a qPCR Ct (cycle threshold) value of 36) was clearly positive (Fig. 5e,f). We also combined sPAMC with lateral-flow assay strips, and this combination was able to detect viral samples with Ct values of 33–34 (Fig. 5g). These results are consistent with previous studies showing that lateral-flow strip assays are less sensitive than their corresponding fluorescence signal-based assays22,33. The readout by lateral flow requires opening of tubes to add buffer and a strip, an additional step that increases hands-on and waiting times, as well as risk of cross-contamination22,33. As fluorescence readout stimulated by simple UV light is easier, faster and more sensitive, we decided to use it for sPAMC to detect SARS-CoV-2 in samples.
a–d, The sensitivity of suboptimal PAM-mediated one-pot reaction (b,d) and qPCR assay (a,c) targeting the UL55 gene of HCMV was compared. The PUC57-UL55 plasmid was used as substrate in a and b. The presence of HCMV virus was determined in c and d. The reaction volume of qPCR in a and c is 20 μl and the reaction volume of one-pot reactions in b and d is 30 μl. The numbers of copies inputted in the two reactions were the same. e, Schematic of detection under portable UV light and using a lateral flow strip. The lysed viral samples were used as an input to sPAMC, which was visualized by UV light or lateral flow strip. f, The direct fluorescence stimulated by UV light was visualized to detect HCMV virus. The reaction was examined under UV light at 8, 10, 15 and 20 min after incubation at 37 °C. g, At 20 min after incubation, a lateral flow strip was dipped into the reaction tube for 5 min to visualize the control and test bands. Red arrows indicate the flow direction of samples in strips.
One strength of sPAMC is that it greatly expands the available selection of crRNAs as there are more suboptimal PAMs than canonical PAMs. Spacers using VTTV, TCTV and TTVV PAMs likely perform well in the one-pot reaction, making the number of available suboptimal PAMs 7-fold higher than that of canonical PAMs in theory (21 combinations versus 3 combinations) (Fig. 6a and Supplementary Fig. 11a). Moreover, some additional suboptimal PAMs, such as TRTV, TTNT and YYYN (except TTTV) may also function better than canonical PAMs, making the choice of spacer even more flexible (Supplementary Fig. 11b,c). The relaxed criteria of PAM selection are particularly important for developing test kits for viral detection. Although there are more than 1,000 canonical PAMs of Cas12a in SARS-CoV-2, only a limited number of canonical PAMs could be employed for viral detection assays given the selection criteria: (1) in a conserved region, (2) in a high-copy gene, (3) an active crRNA and (4) compatible with robust primers for isothermal amplification. Hence, the extended selection of suboptimal PAMs makes sPAMC more flexible for assay optimization and application to different viral strains.
a, Numbers of canonical PAM (TTTV) and suboptimal PAM (VTTV, TTVV, TCTV) spacers in SARS-CoV-2. b–e, Detection limits of sPAMC (b,d) at 42 °C and STOPCovid.v1 (c,e) at 60 °C on DNA (left) and RNA (right). Fluorescence values at 15 min in sPAMC and 60 min in STOPCovid.v1 are shown in b and c, respectively, and the relevant fluorescence curves were shown in d and e. The numbers of molecules inputted in sPAMC and STOPCovid.v1 were the same. Mean ± s.d. of n = 3 technical replicates for b and c. f, sPAMC results for 204 SARS-CoV-2 nasopharyngeal swab samples (left, 104 positive samples, 48 unextracted samples marked by solid circle and 56 pre-extracted samples marked by hollow circle; right, 100 negative samples). The fluorescence readout was measured at 20 min at 42 °C. The threshold was determined as 3× the average of all samples’ initial fluorescence values, at a signal-to-noise ratio of 3. g, Direct visualization under UV light to detect unextracted SARS-CoV-2 positive samples. The reaction was examined under UV light at 10, 15 and 20 min after incubation at 42 °C. h, Concordance table between sPAMC and RT–qPCR for 204 samples.
Finally, we applied sPAMC to detect SARS-CoV-2. We first compared the sensitivity of RPA/Cas12a/suboptimal PAM-based sPAMC and LAMP/Cas12b-based STOPCovid using a DNA fragment encoded nucleoprotein (N) gene of SARS-CoV-2. sPAMC is ~100-fold more sensitive than STOPCovid, and the speed of sPAMC is 3-fold faster than STOPCovid in detecting DNA samples (13 min versus 40 min, time to half-maximum fluorescence) (Fig. 6b–e, left). Then we examined the capability of sPAMC to detect RNA samples by combining RT–RPA and Cas12a. Our initial data showed that the sPAMC was not as sensitive as RT–qPCR in detecting RNA samples. We speculated that the RT step was the rate-limiting step, despite RNase H having been added to the reaction33,37. To improve the efficiency of the RT step, we first increased the temperature of the reaction from 37 °C to 42 °C, as RT enzymes usually perform better at higher temperature, and both RPA and Cas12a are activated at 42 °C. Indeed, sPAMC performed well at 42 °C (Supplementary Fig. 12a). Similar to RT–qPCR, RT–RPA usually used its reverse primer as RT primer33,37. We hypothesized that the RPA reverse primer which is more than 30 nt, may not be efficient for the RT step. Indeed, the RT efficiency of the RPA reverse primer was 6-fold less efficient than the qPCR reverse primer (Supplementary Fig. 12b). Therefore, we added an additional 18 nt primer to function as an RT primer, and meanwhile reduced the concentration of the RPA reverse primer to prevent its interference of the RT process. The combination of reactions at 42 °C, adding an additional short RT primer and reducing the concentration of RPA reverse primer substantially improved RT efficiency and overall sPAMC performance for detecting RNA samples (Supplementary Fig. 12a–d). An in vitro-transcribed RNA fragment of SARS-CoV-2 N gene was used to compare the ability of sPAMC and STOPCovid to detect RNA. sPAMC exhibited ~100-fold higher sensitivity, and was ~2.5-fold faster than STOPCovid for RNA detection (Fig. 6b–e, right).
We head-to-head compared the limits of detection (LOD) of sPAMC and RT–qPCR, the latter considered as the current gold standard by US Centres for Disease Control and Prevention (CDC)38. We first determined the LOD of RT–qPCR using commercially available SARS-CoV-2 Pseudovirus as the standard37. The LOD of RT–qPCR was 1 cp μl−1 (Supplementary Fig. 13a,b), which is consistent with the results released by CDC38. Then we compared the LOD of sPAMC and RT–qPCR, and determined that the LOD of sPAMC was 1 cp μl−1 (Supplementary Fig. 13c), comparable to that of RT–qPCR.
We evaluated the performance of sPAMC in SARS-CoV-2-positive and -negative clinical samples, and compared its performance with STOPCovid. A total of 104 SARS-CoV-2-positive nasopharyngeal swab samples (48 unextracted and 56 extracted samples) with a wide range of Ct values (18.1–35.8) and 100 SARS-CoV-2-negative samples were used. sPAMC had a sensitivity of 94.2% and a specificity of 100.0% (Fig. 6f–h), and it was able to detect samples with Ct value 35.8 (1.2 cp μl−1 according to the standard curve of RT–qPCR in Supplementary Fig. 13b). The positive signal appeared as early as 10 min, and all positive samples showed signals at 15 min (Fig. 6g and Supplementary Fig. 14a,b). The signal can be detected by UV light or a simple blue light device (Supplementary Fig. 14c). In comparison, STOPCovid.v1 was unable to stably detect samples with Ct value above 31.0, resulting in a sensitivity of 78.8% (Supplementary Fig. 15a–c). Finally, to assess the specificity of sPAMC, we tested several common human coronaviruses including MERS, HKU1, 229E, NL63 and OC43 through RPA amplification, collateral activity test and one-pot reaction. The results indicated no cross-reaction with other common viruses (Supplementary Fig. 16a–d).
Discussion
The global outbreak of COVID-19 highlights the critical need for fast, sensitive and reliable diagnostics that can be applied in the laboratory and for point-of-care testing13,14,15,16,17,18,19,20. RT–qPCR is the most widely used gold-standard test for COVID-19. However, its relatively long turnaround time and reliance on expensive equipment have become rate-limiting issues when facing high demands for virus tests13,14,15,16. To meet these great challenges and help stop the spread of COVID-19, a preferable molecular assay fulfilling the following criteria is greatly needed to monitor and manage viral spread: (1) fast, to provide diagnostic information within the period of a patient visit or before a gathering event, (2) simple, to allow the test to be used by unskilled people and (3) equally sensitive as RT–qPCR, to capture most infected patients.
Here we developed a suboptimal PAM-mediated one-pot detection method named ‘sPAMC’. We found that using a suboptimal PAM of Cas12a rather than canonical PAM provides robust one-pot test in several ways: (1) the speed of sPAMC is 2- to 3-fold faster than that of a one-pot reaction using canonical PAM (Fig. 1, Extended Data Figs. 1–3, and Supplementary Figs. 1–4 and 6), requiring only 10–15 min to detect HCMV and 15 min to detect SARS-CoV-2 (Figs. 5 and 6); (2) the sensitivity of sPAMC is higher than that of a one-pot test using canonical PAM, and comparable to that of qPCR (Figs. 2, 5 and 6); (3) the signal from sPAMC is highly reproducible, with less than ~30% fluctuation across samples (Fig. 2); and (4) the broader selection of suboptimal PAM sequences makes assay development more flexible (Fig. 6a and Supplementary Fig. 11). Mechanistically, we found that Cas12a-mediated substrate cleavage competes with isothermal amplification in the one-pot reaction. The reduced binding affinity of Cas12a with suboptimal PAM substrate is critical for the attenuated cis-cleavage activity, and thus promotes the balance shift to isothermal amplification (Figs. 1, 3 and 4, Supplementary Table 1, Extended Data Fig. 2, and Supplementary Figs. 1–4 and 6). As a consequence, effective isothermal amplification further strengthens Cas12a-mediated detection by providing a sufficient pool of amplicons within a short period of time, allowing the detection of low concentrations of substrates and the generation of reproducible and robust signals.
A kinetics model based on Michaelis–Menten enzyme kinetics theory was developed for Cas12 and Cas1339. It indicates that cis cleavage retains sufficient activities even at low substrate concentrations, and increasing the concentrations of substrates accelerates collateral reaction39. This is consistent with our model that cis cleavage is the key to the performance of one-pot reactions (Figs. 3 and 4, and Supplementary Table 1). Due to excessive substrate consumption caused by canonical PAMs-mediated cis cleavage at low concentration of substrates, amplicon accumulation is slow and unstable, resulting in delayed or lack of collateral activity. In contrast, suboptimal PAMs allow substrate accumulation at early stages of the isothermal reaction, and the abundant amplicons further accelerate collateral reaction, resulting in faster reaction with even lower concentrations of initial substrate.
The sequencing ratio stabilizes within the first minute in Fig. 3c,d. The rapid stabilization of the sequencing ratio can be caused by stronger cis-cleavage activity of canonical PAMs to quickly reduce their substrates at very early time points (within the first minute of reaction). This is consistent with the cis-cleavage data of canonical PAMs and suboptimal PAMs (cleavage data at 0.5 and 1 min in the gel; Fig. 3e,f and Supplementary Fig. 9).
We applied several methods to detect unextracted NP swabs, including fluorescence readout, UV imaging and blue light imaging by simple devices (Fig. 6f,g and Supplementary Fig. 14). Both UV and blue light can effectively detect all positive samples. Strong positive samples could be visualized with naked eyes. Further optimizing fluorescence reporters or improving collateral activity may boost the signal to allow all positive samples to be visualized with naked eyes36.
A recent study clinically validated a two-step method for Cas13a-mediated detection of SARS-CoV-233. The two-step detection took approximately 1 h, and its detection limit corresponded to a Ct value of 33.5 in RT–qPCR33. Cas12a-based DETECTR was applied to detect SARS-CoV-2 from RNA extracts of patient samples, and it demonstrated 95% positive agreement with RT–qPCR assay32. DETECTR is also a two-step detection method involving RT-LAMP and subsequent Cas12a detection, taking approximately 1 h to finish32. In comparison, the sPAMC developed in this study is easier to perform, following a one-step protocol to minimize hands-on time and incubation time, and reduce the probability of contamination.
STOPCovid.v1 combines LAMP and Cas12b detection in one step22. A magnetic bead step for RNA extraction and concentration is added before the one-pot reaction to generate STOPCovid, version 2 (STOPCovid.v2)22. STOPCovid.v1 (200 copies per reaction) is less sensitive than RT–qPCR (20 copies per reaction), while STOPCovid.v2’s detection limit has been substantially improved due to the RNA extraction and concentration step22. Although RNA extraction is able to enhance sensitivity, the extra step of using magnetic beads and fields increases the test cost and hands-on time, and an entire nasopharyngeal swab is used for one test, making it difficult to repeat the assay. In both versions, Cas12b uses its canonical PAM22. It is possible that suboptimal PAMs might function similarly in promoting Cas12b-based one-pot reaction. The notable variation in the fluorescence signal in the STOP assay might be minimized by using suboptimal PAM22. The spacer used in STOPCovid partially overlaps with the primers used for LAMP; thus, the concentrations of Cas12b and crRNA are titrated to reduce false-positive results22. The limited choice of spacers due to the lower prevalence of canonical PAMs may introduce such overlap. Targeting broadly available suboptimal PAMs may avoid the need to overlap the spacer with LAMP primers and also allow the identification of better-performing spacers to further improve the performance of STOPCovid.
The approaches of using suboptimal PAM or reducing binding of Cas effectors to their substrates in a one-pot reaction could also be tested for other members of the Cas12a family and Class II type V effectors. It will be interesting to explore whether Cas proteins other than Cas12a could exhibit superior speed and sensitivity using suboptimal PAM in a one-pot reaction. Although there remain several future directions to pursue, sPAMC requires less incubation time and is more sensitive than STOPCovid.v1, SHINE and amplification-free detection using Cas13a (Extended Data Fig. 4). Moreover, it is easier to perform and faster than SHERLOCK, DETECTR and STOPCovid.v2. sPAMC appears to be the first CRISPR-mediated detection method to exhibit the combination of one step, fast speed, high sensitivity, high reliability and flexibility.
Methods
Plasmid and dsDNA preparation
The S and E genes of SARS-CoV-2 were synthesized and cloned into the pUC57 vector (GenScript Biotech). The N gene dsDNA of SARS-CoV-2 was obtained by RT–PCR using inactivated viruses, and N gene dsDNA of other human coronaviruses were synthesized (GenScript). The Orf1ab dsDNA substrates containing spacer 4 and spacer 5 targeting regions were obtained by PCR. UL55 dsDNA was obtained by PCR using inactivated HCMV virus as a template and cloned into the pUC57 vector. The SARS-CoV-2 Pseudovirus was lentivirus packaged with SARS-CoV-2 N gene (Beyotime Biotechnology).
Preparation of HCMV
Viral samples were collected from the supernatant of cells cultured after infection with HCMV. The HCMV viral sample was inactivated at 95 °C and diluted 1:1 in lysis buffer (QuickExtract DNA extraction solution). The copy number was quantified by qPCR according to the standard curve generated using plasmid DNA.
LbCas12a protein expression and purification
The DNA fragment encoding LbCas12a was cloned into a pET-based expression vector containing a C-terminal 6×His-tag. Escherichia coli strain BL21 (DE3) transformed with the recombinant plasmid was incubated with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the culture density reached an OD600 of 0.7 and was grown at 21 °C for another 16 h. The protein was purified from the cell lysate via Ni-NTA resin and eluted with buffer containing 20 mM Tris-HCl, 500 mM NaCl and 500 mM imidazole, pH 7.4. Then, the concentrated protein was further filtered using a gel filtration column (Superdex 200 Increase 10/300 GL) in elution buffer containing 20 mM Tris-HCl and 200 mM NaCl at pH 7.5. The final storage buffer comprised 20 mM Tris-HCl (pH 7.5), 200 mM NaCl and 5% glycerol.
RPA and RT–RPA
One lyophilised RPA pellet was resuspended in 29.4 μl buffer A, 16.1 μl nuclease-free water, 1 μl 20 μM RPA forward primer and 1 μl 20 μM RPA reverse primer to form the RPA mix according to the manufacturer’s instructions (Weifang Amp-Future Biotech). RPA kits from TwistDx (TABAS03KIT) were used in Supplementary Fig. 10. TwistDx RPA mix was resuspended in 29.5 μl rehydration buffer, 15.6 μl nuclease-free water, 1.2 μl 20 μM RPA forward primer and 1.2 μl 20 μM RPA reverse primer. The primer sequences are presented in Supplementary Table 2. For the RT–RPA reaction, 0.9 μl RNase H (50 U μl−1 stock; New England Biolabs, NEB) and 0.45 μl SuperScript IV reverse transcriptase (Thermo Fisher) or EpiScript RNase H-reverse transcriptase (Lucigen) were added to the RPA mixture33,37. The reactions were performed at 37 °C or 42 °C. In the finalized version of RT–RPA reactions, 1 μl 5 μM RPA reverse primer and an additional RT primer (1 μl 40 μM) were added.
Preparation of crRNA
The DNA template for in vitro transcription was synthesized by overlapping PCR of two oligos. One oligo contained the T7 promoter sequence, and the other contained a spacer sequence. The PCR product was incubated with T7 RNA polymerase for in vitro transcription at 37 °C for 2 h. The in vitro transcription reaction was treated with DNase I (Promega) for 15 min at 37 °C, and then purified using a Monarch RNA cleanup kit (NEB). The crRNA sequences are presented in Extended Data Fig. 1c and Supplementary Table 3.
One-pot reaction
One-pot reactions were performed in 30 μl reaction volumes containing 33 or 100 nM LbCas12a RNP, 400 nM FQ ssDNA reporter (FAM-TTATT-Quencher, Takara Biotechnology), dsDNA substrate (Supplementary Table 4) and RPA or RT–RPA components. The RNP complex, FQ ssDNA reporter (8 μl) and RPA mixture (18 μl) were added to each one-pot reaction well, and subsequently, 2 μl buffer B and dsDNA activator were supplied before readout through a SpectraMax i3x at 37 °C or 42 °C. The assay was also monitored under UV, blue light or by lateral flow detection (Milenia HybriDetect 1 kit, TwistDx). The final concentrations of the reporter for UV detection were adjusted to 0.4–2 μM. The reporter for lateral flow detection was FAM-TTATTATT-Biotin, with a final concentration of 0.8 μM. The concentrations of dsDNA substrate were 18.3 fM–2.3 pM for Fig. 1, and Supplementary Figs. 1–4 and 6.
Deep sequencing
The deep-sequencing samples were prepared as one-pot reactions, except that substrate consisted of mixed canonical-PAM and suboptimal-PAM substrates at a 1:1 ratio. The reaction was terminated by adding proteinase K (Thermo Fisher) at different time points, and then heated at 95 °C for 5 min to inactivate the protease. The products were amplified with adapters and barcodes (Supplementary Table 5) for Illumina NovaSeq, and the reads were filtered using an average Phred quality (Q score) of at least 25. Raw reads were analysed by Python scripts, and the data were normalized according to the reads at the 0 min time point.
Cas12a in vitro cleavage and collateral activity
For in vitro cleavage, the LbCas12a RNP was incubated at room temperature for 20 min in 1× NEBuffer 2.1 before incubation with dsDNA at 37 °C. The reaction was terminated by adding proteinase K at various time points, and the products were visualized on a 2% Tris-acetate-EDTA (TAE) gel. The concentrations of RNP used were 50 or 100 nM, and the concentrations of dsDNA substrate were 6–7.5 nM or 9–11 nM. The percentage of substrates and products were quantified by Image Lab software (Bio-Rad). The cleavage efficiency at each time point was plotted as a function of time, and these data were fit with a one-phase exponential decay curve, to calculate Kcleave values (Prism 8, GraphPad Software)40. The collateral activity assay was performed in a 30 μl volume containing 33 nM LbCas12a RNP and 400 nM ssDNA reporter (FAM-TTATT-BHQ1) in 1× NEBuffer 2.1, and the fluorescence signal was recorded using SpectraMax i3x. The concentrations of dsDNA substrate activators used were 2.7–3.5 nM.
EMSA
Deactivated LbCas12a (D832A) (hereafter dCas12a) was expressed and purified as described above. An EMSA was performed with dCas12a RNP and a 5’-FAM-labelled 50 nt dsDNA substrate using 1× NEBuffer 2.1. Binding was carried out at 37 °C for 20 min and the reactions were then supplemented with 5% glycerol. Samples were then resolved on 4% Tris-borate/EDTA polyacrylamide gels for 15–20 min at 120 V, and the results were visualized using a fluorescent image analyser.
qPCR and RT–qPCR assay
qPCR assays for HCMV samples were performed in 20 μl reaction volumes containing 10 μl 2× AceQ qPCR probe master mix (Vazyme), 1 μl of each primer pair at 10 μM (Supplementary Table 6) and 0.2 μl 10 μM TaqMan probe (GenScript). The numbers of viral copies inputted and sample processing in qPCR and sPAMC were the same. Each RT–qPCR reaction for SARS-CoV-2 samples contained 10 μl 2× one-step SYBR Green mix, 1 μl one-step SYBR Green enzyme mix (Vazyme) and 0.4 μl of the primer pairs at 10 μM. The input volume for the RT–qPCR assay was 1.34 μl sample per 20 μl reaction.
SARS-CoV-2 clinical sample collection
The clinical samples tested in this study were approved by the Wuhan Jinyintan Hospital Ethics Committee (KY-2021-01.01). SARS-CoV-2 samples were obtained from the Wuhan Jinyintan Hospital. Unextracted samples were lysed at 95 °C for 5–10 min with an equal volume of lysis buffer containing 1 U μl−1 RNasin Plus, 250 μM tris-2-chloroethyl phosphate (TCEP) and 0.02 μg μl−1 Chelex-10037,41. The extracted RNA samples were purified according to the manufacturer’s protocol (liferiver). They were mixed with RT primer for sPMAC detection. The UV images for all samples were processed in Image Lab (Bio-Rad) under these parameters: time of exposure: 0.368–0.636 s, Gamma value: 0.9–1.14. STOPCovid.v1 assay was performed exactly following the published protocol22.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data supporting the results in this study are available within the paper and its Supplementary Information. The deep-sequencing raw data are available from the Sequence Read Archive (SRA) of NCBI under accession number PRJNA728523. Source data are provided with this paper.
References
Kuiken, T. et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362, 263–270 (2003).
Zhong, N. S. et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362, 1353–1358 (2003).
Zaki, A. M., van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. & Fouchier, R. A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012).
Chang, S. Y., Bowman, B. H., Weiss, J. B., Garcia, R. E. & White, T. J. The origin of HIV-1 isolate HTLV-IIIB. Nature 363, 466–469 (1993).
Dietrich, U. et al. A highly divergent HIV-2-related isolate. Nature 342, 948–950 (1989).
Musso, D., Ko, A. I. & Baud, D. Zika virus infection - after the pandemic. N. Engl. J. Med. 381, 1444–1457 (2019).
Piot, P., Muyembe, J. J. & Edmunds, W. J. Ebola in west Africa: from disease outbreak to humanitarian crisis. Lancet Infect. Dis. 14, 1034–1035 (2014).
Wang, C., Horby, P. W., Hayden, F. G. & Gao, G. F. A novel coronavirus outbreak of global health concern. Lancet 395, 470–473 (2020).
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Rothe, C. et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382, 970–971 (2020).
Bai, Y. et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA 323, 1406–1407 (2020).
Harrison, A. G., Lin, T. & Wang, P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 41, 1100–1115 (2020).
Vogels, C. B. F. et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer-probe sets. Nat. Microbiol. 5, 1299–1305 (2020).
Osório, N. S. & Correia-Neves, M. Implication of SARS-CoV-2 evolution in the sensitivity of RT–qPCR diagnostic assays. Lancet Infect. Dis. 21, 166–167 (2021).
Vanaerschot, M. et al. Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by RT-PCR. J. Clin. Microbiol. 59, e02369 (2020).
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020).
Piepenburg, O., Williams, C. H., Stemple, D. L. & Armes, N. A. DNA detection using recombination proteins. PLoS Biol. 4, e204 (2006).
Zaghloul, H. & El-Shahat, M. Recombinase polymerase amplification as a promising tool in hepatitis C virus diagnosis. World J. Hepatol. 6, 916–922 (2014).
Park, G. S. et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 22, 729–735 (2020).
Baek, Y. H. et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg. Microbes Infect. 9, 998–1007 (2020).
Basu, A. et al. Performance of Abbott ID Now COVID-19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City academic institution. J. Clin. Microbiol. 58, e01136-20 (2020).
Joung, J. et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 383, 1492–1494 (2020).
Arizti-Sanz, J. et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 11, 5921 (2020).
Wu, H. et al. A recombinase polymerase amplification and lateral flow strip combined method that detects salmonella enterica serotype typhimurium with no worry of primer-dependent artifacts. Front. Microbiol. 11, 1015 (2020).
Hardinge, P. & Murray, J. A. H. Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci. Rep. 9, 7400 (2019).
Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Teng, F. et al. CDetection: CRISPR–Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 20, 132 (2019).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Li, S.-Y. et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491–493 (2018).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356, 438–442 (2017).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR–Cas13. Science 360, 444–448 (2018).
Broughton, J. P. et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874 (2020).
Patchsung, M. et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 4, 1140–1149 (2020).
Fozouni, P. et al. Amplification-free detection of SARS-CoV-2 with CRISPR–Cas13a and mobile phone microscopy. Cell 184, 323–333 (2021).
Yamano, T. et al. Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR–Cpf1. Mol. Cell 67, 633–645.e3 (2017).
Ding, X. et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR–Cas12a assay. Nat. Commun. 11, 4711 (2020).
Qian, J. et al. An enhanced isothermal amplification assay for viral detection. Nat. Commun. 11, 5920 (2020).
Real-time RT–PCR Panel for Detection of 2019-nCoV (US Centers for Disease Control and Prevention, 2020); https://www.fda.gov/media/134922/download
Ramachandran, A. & Santiago, J. G. CRISPR enzyme kinetics for molecular diagnostics. Anal. Chem. 93, 7456–7464 (2021).
Ma, E., Harrington, L. B., O’Connell, M. R., Zhou, K. & Doudna, J. A. Single-stranded DNA cleavage by divergent CRISPR–Cas9 enzymes. Mol. Cell 60, 398–407 (2015).
Lee, R. A. et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl Acad. Sci. USA 117, 25722–25731 (2020).
Acknowledgements
This work was kindly supported by the National Key R&D Program of China (2019YFA0802801 and 2018YFA0801401), the Strategic Priority Research Program of CAS (XDB29010300 to X.Z.), the National Natural Science Foundation of China (31871345 and 32071442 to H.Y., 31972936 to Ying Zhang, 31970169 to X.Z., 32000115 to Y.H.), the Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (TFJC2018004), the Fundamental Research Funds for the Central Universities (to H.Y. and Ying Zhang), the Applied Basic Frontier Program of Wuhan City (2020020601012216 to H.Y.), the Hubei Health Commission Young Investigator award (to H.Y.) the startup funding from Wuhan University (to H.Y and Ying Zhang), and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2020-PT320-004). We thank the core facility of the Medical Research Institute at Wuhan University for their technical support.
Author information
Authors and Affiliations
Contributions
H.Y. conceived, designed and managed the project; S.L. and X.T. performed most experiments with the help of K.Z., Q.C., J.D. and X.L.; J.D. and Yizhou Zhang performed bioinformatics analyses; Ying Zhang provided conceptual advice; D.-Y.Z, X.Z., Y.H., Y.Q. and M.H. provided viral samples and performed clinical samples related experiments; H.Y., Ying Zhang and X.Z. analysed the data; H.Y. and Ying Zhang wrote the paper with inputs from the other authors.
Corresponding authors
Ethics declarations
Competing interests
H.Y., S.L., Ying Zhang, X.T., K.Z., X.Z. and D.-Y.Z. have a filed patent application on sPAMC through Wuhan University (PCT International Application No. PCT/CN2022/070850, filing date 7 January 2022). The other authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Tony Hu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Suboptimal and canonical PAMs-mediated one-pot reactions.
a,b, One-pot reactions used spacers with suboptimal or canonical PAMs in Orf1ab (a) and E (b) genes of SARS-CoV-2. The crRNAs 1–3 targeting Orf1ab gene and crRNAs 2–7 targeting E gene used canonical PAMs whereas crRNAs 4-5 targeting Orf1ab gene and crRNA 1 targeting E gene used suboptimal PAMs. c, The PAMs and spacers used in the one-pot reaction.
Extended Data Fig. 2 Comparison of collateral activity and one-pot reaction on various suboptimal and canonical PAMs.
a-d, Summary of fluorescent kinetics for position 1–3 point-mutated suboptimal PAMs and three canonical PAMs in collateral activity test (a-b) and the corresponding one-pot reaction of HPV18 L1 gene spacer 1 (c) and SARS-CoV-2 S gene spacer 2 (d). Time to half-maximum fluorescence was determined. Fluorescence values were determined at 40 and 20 minute for collateral activities and one-pot reactions, respectively. 2.7 nM and 2.8 nM dsDNA substrates were used for collateral activity assays of HPV18 L1 gene spacer 1 and S gene spacer 2, respectively. 18.3 fM and 189 fM dsDNA substrates were used for one-pot reaction of HPV18 L1 gene spacer 1 and S gene spacer 2, respectively. Mean ± s.d. of 3 technical replicates.
Extended Data Fig. 3 Comparison of suboptimal and canonical PAM-mediated one-pot reaction.
E gene spacer 8 (a) and S gene spacer 3 (b) of SARS-CoV-2 were examined. The concentrations of dsDNA in one-pot reactions were 325.5 fM and 189 fM for E gene spacer 8 and S gene spacer 3, respectively. All reactions were carried out at 37 °C.
Extended Data Fig. 4 Comparison of CRISPR-based SARS-CoV-2 detection methods.
The substrates for evaluating sensitivity are the following: SARS-CoV-2 Pseudovirus for sPAMC, N gene RNA for DETECTR and Amplification-free detection, extracted genomic RNA for SHERLOCK and SHINE, SARS-CoV-2 genome standards for STOPCovid.v1, and concentrated samples for STOPCovid.v2.
Supplementary information
Supplementary Information
Supplementary tables and figures.
Supplementary Data 1
Summary of ranked PAM by one-pot reaction performance and cis-cleavage activities.
Supplementary Data 2
Data for Supplementary Fig. 1.
Supplementary Data 3
Data for Supplementary Fig. 2.
Supplementary Data 4
Data for Supplementary Fig. 3.
Supplementary Data 5
Data for Supplementary Fig. 4.
Supplementary Data 6
Data for Supplementary Fig. 6.
Supplementary Data 7
Data for Supplementary Fig. 7.
Supplementary Data 8
Data for Supplementary Fig. 10.
Supplementary Data 9
Data for Supplementary Fig. 12.
Supplementary Data 10
Data for Supplementary Fig. 13.
Supplementary Data 11
Data for Supplementary Fig. 15.
Supplementary Data 12
Data for Supplementary Fig. 16.
Source data
Source Data Fig. 1
Source data.
Source Data Fig. 2
Source data.
Source Data Fig. 3
Source data.
Source Data Fig. 4
Source data.
Source Data Fig. 5
Source data.
Source Data Fig. 6
Source data.
Source Data Extended Data Fig. 1
Source data.
Source Data Extended Data Fig. 2
Source data.
Source Data Extended Data Fig. 3
Source data.
Rights and permissions
About this article
Cite this article
Lu, S., Tong, X., Han, Y. et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng 6, 286–297 (2022). https://doi.org/10.1038/s41551-022-00861-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-022-00861-x
This article is cited by
-
CRISPR-powered quantitative keyword search engine in DNA data storage
Nature Communications (2024)
-
Nanotechnology’s frontier in combatting infectious and inflammatory diseases: prevention and treatment
Signal Transduction and Targeted Therapy (2024)
-
Fast and sensitive CRISPR detection by minimized interference of target amplification
Nature Chemical Biology (2024)
-
Adeno-associated virus genome quantification with amplification-free CRISPR-Cas12a
Gene Therapy (2024)
-
Ultrasensitive single-step CRISPR detection of monkeypox virus in minutes with a vest-pocket diagnostic device
Nature Communications (2024)