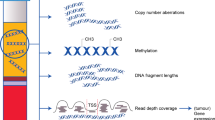
Abstract
Cell-free DNA (cfDNA) in the circulating blood plasma of patients with cancer contains tumour-derived DNA sequences that can serve as biomarkers for guiding therapy, for the monitoring of drug resistance, and for the early detection of cancers. However, the analysis of cfDNA for clinical diagnostic applications remains challenging because of the low concentrations of cfDNA, and because cfDNA is fragmented into short lengths and is susceptible to chemical damage. Barcodes of unique molecular identifiers have been implemented to overcome the intrinsic errors of next-generation sequencing, which is the prevailing method for highly multiplexed cfDNA analysis. However, a number of methodological and pre-analytical factors limit the clinical sensitivity of the cfDNA-based detection of cancers from liquid biopsies. In this Review, we describe the state-of-the-art technologies for cfDNA analysis, with emphasis on multiplexing strategies, and discuss outstanding biological and technical challenges that, if addressed, would substantially improve cancer diagnostics and patient care.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lo, Y. D. et al. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 64, 218–224 (1999).
Diehl, F. et al. Circulating mutant DNA to assess tumour dynamics. Nat. Med. 14, 985–990 (2008).
Jahr, S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001).
Wan, J. C. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat. Rev. Cancer 17, 223–238 (2017).
Diaz, L. A. Jr & Bardelli, A. Liquid biopsies: genotyping circulating tumour DNA. J. Clin. Oncol. 32, 579–586 (2014).
Thierry, A. R. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumour DNA. Nat. Med. 20, 430–435 (2014).
Alix-Panabieres, C. & Pantel, K. Clinical applications of circulating tumour cells and circulating tumour DNA as liquid biopsy. Cancer Discov. 6, 479–491 (2016).
Mok, T. et al. Detection and dynamic changes of EGFR mutations from circulating tumour DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin. Cancer Res. 21, 3196–3203 (2015).
Hao, T. B. et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br. J. Cancer 111, 1482–1489 (2014).
Azad, A. A. et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin. Cancer Res. 21, 2315–2324 (2015).
Lebofsky, R. et al. Circulating tumour DNA as a non-invasive substitute to metastasis biopsy for tumour genotyping and personalized medicine in a prospective trial across all tumour types. Mol. Oncol. 9, 783–790 (2015).
Schwarzenbach, H., Hoon, D. S. B. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011).
Goyal, L. et al. Polyclonal secondary FGFR2 mutations drive acquired resistance to FGFR inhibition in patients with FGFR2 fusion-positive cholangiocarcinoma. Cancer Discov. 7, 252–263 (2016).
Distribution of In Vitro Diagnostic Products Labeled for Research Use Only or Investigational Use Only: Guidance for Industry and Food and Drug Administration Staff (US FDA, 2013); https://www.fda.gov/regulatory-information/search-fda-guidance-documents/distribution-vitro-diagnostic-productslabeled-research-use-only-or-investigational-use-only
Dietel, M. et al. Diagnostic procedures for non-small-cell lung cancer (NSCLC): recommendations of the European Expert Group. Thorax 71, 177–184 (2015).
Herbst, R. S. et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387, 1540–1550 (2016).
Vansteenkiste, J. et al. Final results of a multi-center, double-blind, randomized, placebo-controlled phase II study to assess the efficacy of MAGE-A3 immunotherapeutic as adjuvant therapy in stage IB/II non-small cell lung cancer (NSCLC). J. Clin. Oncol. 25, 7554 (2007).
Gettinger, S. N. et al. Overall survival and long-term safety of nivolumab (anti-programmed death 1 antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non-small-cell lung cancer. J. Clin. Oncol. 33, 2004–2012 (2015).
Burstein, H. J. et al. Clinical cancer advances 2017: annual report on progress against cancer from the American Society of Clinical Oncology. J. Clin. Oncol. 35, 1341–1367 (2017).
Wender, R. et al. American Cancer Society lung cancer screening guidelines. CA Cancer J. Clin. 63, 107–117 (2013).
Baldwin, D. R. et al. UK Lung Screen (UKLS) nodule management protocol: modelling of a single screen randomised controlled trial of low-dose CT screening for lung cancer. Thorax 66, 308–313 (2011).
Field, J. K. et al. International association for the study of lung cancer computed tomography screening workshop 2011 report. J. Thorac. Oncol. 7, 10–19 (2012).
Spigel, D. R. et al. Final efficacy results from OAM4558g, a randomized phase II study evaluating MetMAb or placebo in combination with erlotinib in advanced NSCLC. J. Clin. Oncol. 29, 7505 (2011).
Mok, T. S. et al. Osimertinib or platinum-pemetrexed in EGFR T790M positive lung cancer. N. Engl. J. Med. 376, 629–640 (2017).
Zhou, C. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Lancet Oncol. 12, 735–742 (2011).
Gatzemeier, U. et al. Results of a phase III trial of erlotinib (OSI-774) combined with cisplatin and gemcitabine (GC) chemotherapy in advanced non-small-cell lung cancer (NSCLC). J. Clin. Oncol. 22, 7010 (2004).
Oxnard, G. R. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 34, 3375–3382 (2016).
Clark, T. A. et al. Analytical validation of a hybrid capture-based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumour DNA. The. J. Mol. Diagnostics 20, 686–702 (2018).
Lanman, R. B. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumour DNA. PLoS ONE 10, e0140712 (2015).
Paz-Ares, L. et al. plus chemotherapy for squamous non–small-cell lung cancer. N. Engl. J. Med. 379, 2040–2051 (2018).
Fabrizio, D. et al. A blood-based next-generation sequencing assay to determine tumour mutational burden (bTMB) is associated with benefit to an anti-PD-L1 inhibitor, atezolizumab. Cancer Res. 78, 5706–5706 (2018).
Hellmann, M. D. et al. (2018). Nivolumab plus ipilimumab in lung cancer with a high tumour mutational burden. N. Engl. J. Med. 378, 2093–2104 (2018).
Gandara, D. R. et al. Blood-based tumour mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 24, 1441–1448 (2018).
Le, D. T. et al. PD-1 blockade in tumours with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumours to PD-1 blockade. Science 357, 409–413 (2017).
Rolfo, C. et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim. Biophys. Acta 1846, 539–546 (2014).
Xiong, L. et al. Dynamics of EGFR mutations in plasma recapitulates the clinical response to EGFR-TKIs in NSCLC patients. Oncotarget 8, 63846–63856 (2017).
Romano, G. et al. A preexisting rare PIK3CAE545K subpopulation confers clinical resistance to MEK plus CDK4/6 inhibition in NRAS melanoma and is dependent on S6K1 signaling. Cancer Discov. 8, 556–567 (2018).
Goldberg, S. B. et al. Early assessment of lung cancer immunotherapy response via circulating tumour DNA. Clin. Cancer Res. 24, 1872–1880 (2018).
Narayan, A. et al. Ultrasensitive measurement of hotspot mutations in tumour DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 72, 3492–3498 (2012).
Thress, K. S. et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small-cell lung cancer harboring EGFR T790M. Nat. Med. 21, 560–562 (2015).
Kwong, L. N. et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat. Med. 18, 1503–1510 (2012).
Wang, Z. et al. Lung adenocarcinoma harboring EGFR T790M and in trans C797S responds to combination therapy of first-and third-generation EGFR TKIs and shifts allelic configuration at resistance. J. Thorac. Oncol. 12, 1723–1727 (2017).
Corcoran, R. B. & Chabner, B. A. Application of cell-free DNA analysis to cancer treatment. N. Engl. J. Med. 379, 1754–1765 (2018).
Garcia-Murillas, I. et al. Mutation tracking in circulating tumour DNA predicts relapse in early breast cancer. Sci. Transl. Med. 7, 302ra133 (2015).
Dawson, S. J. et al. Analysis of circulating tumour DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Reinert, T. et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131 (2019).
Tie, J. et al. Circulating tumour DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8, 346ra92 (2016).
Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC—challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 15, 577–586 (2018).
Oxnard, G. R. et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 20, 1698–1705 (2014).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumour DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Imperiale, T. F. et al. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 370, 1287–1297 (2014).
Chan, K. A. et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 377, 513–522 (2017).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Lecomte, T. et al. Detection of free circulating tumour associated DNA in plasma of colorectal cancer patients and its association with prognosis. N. Engl. J. Med. 370, 1287–1297 (2014).
Hu, Y. et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin. Cancer Res. 24, 4437–4443 (2018).
Heitzer, E., Haque, I. S., Roberts, C. E. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
Sina, A. A. I. et al. Epigenetically reprogrammed methylation landscape drives the DNA self-assembly and serves as a universal cancer biomarker. Nat. Commun. 9, 4915 (2018).
Lehmann-Werman, R. et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl Acad. Sci. USA 113, E1826–E1834 (2016).
Deng, J. et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 27, 353–360 (2009).
Xu, R. H. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 16, 1155–1161 (2017).
Teschendorff, A. E. & Relton, C. L. Statistical and integrative system-level analysis of DNA methylation data. Nat. Rev. Genet. 19, 129–147 (2018).
Guo, S. et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumour tissue-of-origin mapping from plasma DNA. Nat. Genet. 49, 635–642 (2017).
Tug, S. et al. Exercise-induced increases in cell-free DNA in human plasma originate predominantly from cells of the haematopoietic lineage. Exerc. Immunol. Rev. 21, 164–173 (2015).
Moreira, V. G., Prieto, B., Rodr.ıguez, J. S. M. & Alvarez, F. V. Usefulness of cell-free plasma DNA, procalcitonin and C-reactive protein as markers of infection in febrile patients. Ann. Clin. Biochem. 47, 253–258 (2010).
Burnham, P. et al. Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat. Commun. 9, 2412 (2018).
Siljan, W. W. et al. Circulating cell-free DNA is elevated in community acquired bacterial pneumonia and predicts short-term outcome. J. Infect. 73, 383–386 (2016).
Lindahl, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993).
Richter, C., Park, J. W. & Ames, B. N. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc. Natl Acad. Sci. USA 85, 6465–6467 (1988).
Cooke, M. S., Evans, M. D., Dizdaroglu, M. & Lunec, J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17, 1195–1214 (2003).
Norton, S. E., Lechner, J. M., Williams, T. & Fernando, M. R. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin. Biochem. 46, 1561–1565 (2013).
Manning, J. E. in Emergency Medicine: A Comprehensive Study Guide (ed. Tintinalli, J. E.) 227 (McGraw-Hill, 2004).
Botezatu, I. et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin. Chem. 46, 1078–1084 (2000).
Koide, K. et al. Fragmentation of cell-free fetal DNA in plasma and urine of pregnant women. Prenat. Diagnosis 25, 604–607 (2005).
Tani, M. & Beck, S. Epigenome-wide association studies for cancer biomarker discovery in circulating cell-free DNA: technical advances and challenges. Curr. Opin. Genet. Dev. 42, 48–55 (2017).
Reckamp, K. L. et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J. Thorac. Oncol. 11, 1690–1700 (2016).
Swanson, P. et al. Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from London: comparison to VERSANT HIV-1 RNA 3.0, AMPLICOR HIV-1 MONITOR v1. 5, and LCx^A R○ HIV RNA quantitative assays. J. Virol. Methods 137, 184–192 (2006).
Castle, P. E. Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol. 12, 880–890 (2011).
Sandri, M. T. et al. Comparison of the Digene HC2 assay and the Roche AMPLICOR human papillomavirus (HPV) test for detection of high-risk HPV genotypes in cervical samples. J. Clin. Microbiol. 44, 2141–2146 (2006).
Misawa, Y. et al. Application of loop-mediated isothermal amplification technique to rapid and direct detection of methicillin-resistant Staphylococcus aureus (MRSA) in blood cultures. J. Infect. Chemother. 13, 134–140 (2007).
Ethridge, S. F. et al. Performance of the Aptima HIV-1 RNA qualitative assay with 16- and 32-member specimen pools. J. Clin. Microbiol. 48, 3343–3345 (2010).
Pao, W. et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2, e73 (2005).
Mok, T. S. et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N. Engl. J. Med. 376, 629–640 (2017).
Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610 (2011).
Newton, C. R. et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17, 2503–2516 (1989).
Das, J. et al. An electrochemical clamp assay for direct, rapid analysis of circulating nucleic acids in serum. Nat. Chem. 7, 569–575 (2015).
Lin, M. et al. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 11, 1244–1263 (2016).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017).
Dressman, D., Yan, H., Traverso, G., Kinzler, K. W. & Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA 100, 8817–8822 (2003).
Khoo, C. et al. Molecular methods for somatic mutation testing in lung adenocarcinoma: EGFR and beyond. Transl. Lung Cancer Res. 4, 126–141 (2015).
Ragoussis, J. Genotyping technologies for genetic research. Annu. Rev. Genomics Hum. Genet. 10, 117–133 (2009).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Wetterstrand, K. A. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) (National Human Genome Research Institute, accessed 19 July 2018); www.genome.gov/sequencingcostsdata
Laver, T. et al. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 3, 1–8 (2015).
Carneiro, M. O. et al. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics 13, 375 (2012).
Taylor, A. D., Micheel, C. M., Anderson, I. A., Levy, M. A. & Lovly, C. M. The path (way) less traveled: a pathway-oriented approach to providing information about precision cancer medicine on My Cancer Genome. Transl. Oncol. 9, 163–165 (2016).
Diehn, M. et al. Early prediction of clinical outcomes in resected stage II and III colorectal cancer (CRC) through deep sequencing of circulating tumour DNA (ctDNA). J. Clin. Oncol. 35, 3591 (2017).
Couraud, S. et al. Non-invasive diagnosis of actionable mutations by deep sequencing of circulating-free DNA in non-small-cell lung cancer: findings from BioCAST/IFCT-1002. Clin. Cancer Res. 20, 4613–4624 (2014).
Song, P. et al. Selective multiplexed enrichment for the detection and quantitation of low-fraction DNA variants via low depth sequencing. Nat. Biomed. Eng. 5, 690–701 (2021).
Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 (2010).
Hovelson, D. H. et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumours. Neoplasia 17, 385–399 (2015).
Dupuis, J. R. et al. HiMAP: robust phylogenomics from highly multiplexed amplicon sequencing. Mol. Ecol. Resour. 18, 1000–1019 (2018).
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012).
Blakely, C. M. et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat. Genet. 49, 1693–1704 (2017).
Kinde, I. et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. USA 108, 9530–9535 (2011).
Lo, Y. D. et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010).
Thierry, A. R. et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 38, 6159–6175 (2010).
Snyder, M. W., Kircher, M., Hill, A. J., Daza, R. M. & Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164, 57–68 (2016).
Mouliere, F. et al. Enhanced detection of circulating tumour DNA by fragment size analysis. Sci. Transl. Med. 10, 466 (2018).
Thakur, B. K. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 24, 766–769 (2014).
Balaj, L. et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2, 180 (2011).
Zhang, J. X. et al. Predicting DNA hybridization kinetics from sequence. Nat. Chem. 10, 91–98 (2018).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumour DNA. Nat. Biotechnol. 34, 547–555 (2016).
Kou, R. et al. Benefits and challenges with applying unique molecular identifiers in next generation sequencing to detect low frequency mutations. PLoS ONE 11, e0146638 (2016).
Jabara, C. B., Jones, C. D., Roach, J., Anderson, J. A. & Swanstrom, R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl Acad. Sci. USA 108, 20166–20171 (2011).
Abascal, F. et al. Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 (2021).
Cohen, J. D. et al. Detection of low-frequency DNA variants by targeted sequencing of the Watson and Crick strands. Nat. Biotechnol. 39, 1220–1227 (2021).
Pel, J. et al. Nonlinear electrophoretic response yields a unique parameter for separation of biomolecules. Proc. Natl Acad. Sci. USA 106, 14796–14801 (2009).
Kidess, E. et al. Mutation profiling of tumour DNA from plasma and tumour tissue of colorectal cancer patients with a novel, high-sensitivity multiplexed mutation detection platform. Oncotarget 6, 2549–2561 (2015).
Song, C. & Liu et al. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res. 44, e146 (2016).
Seyama, T. et al. A novel blocker-PCR method for detection of rare mutant alleles in the presence of an excess amount of normal DNA. Nucleic Acids Res. 20, 2493–2496 (1992).
Arcila, M., Lau, C., Nafa, K. & Ladanyi, M. Detection of KRAS and BRAF mutations in colorectal carcinoma: roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J. Mol. Diagnostics 13, 64–73 (2011).
Orum, H. et al. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res. 21, 5332–5336 (1993).
Milbury, C. A., Li, J. & Makrigiorgos, G. M. Ice-COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic Acids Res. 39, e2 (2011).
Li, J. et al. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 14, 579–584 (2008).
Zuo, Z. et al. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod. Pathol. 22, 1023–1031 (2009).
Wu, L. R. et al. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat. Biomed. Eng. 1, 714–723 (2017).
Ciriello, G. et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45, 1127–1133 (2013).
Mertens, F., Johansson, B., Fioretos, T. & Mitelman, F. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer 15, 371–381 (2015).
Wang, Q. et al. Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief. Bioinform. 14, 506–519 (2012).
Maher, C. A. et al. Transcriptome sequencing to detect gene fusions in cancer. Nature 458, 97–101 (2009).
Zhao, Q. et al. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc. Natl Acad. Sci. USA 106, 1886–1891 (2009).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Whale, A. S. et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 40, e82 (2012).
Pinkel, D. et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20, 207–211 (1998).
Chiu, R. W. et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. Brit. Med. J. 342, c7401 (2011).
Kops, G. J., Weaver, B. A. & Cleveland, D. W. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5, 773–785 (2004).
Mandel, P. & Metais, P. Les acides nucleiques du plasma sanguin chez la homme. C. R. Seances Soc. Biol. Fil. 142, 241–243 (1948).
Bettegowda, C. et al. Detection of circulating tumour DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 (2014).
Aravanis, A. M., Lee, M. & Klausner, R. D. Next-generation sequencing of circulating tumour DNA for early cancer detection. Cell 168, 571–574 (2017).
Razavi, P. et al. Cell-free DNA (cfDNA) mutations from clonal hematopoiesis: implications for interpretation of liquid biopsy tests. J. Clin. Oncol. 35, 11526 (2017).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Schwarzenbach, H., Nishida, N., Calin, G. A. & Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 11, 145–156 (2014).
Azmi, A. S., Bao, B. & Sarkar, F. H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 32, 623–642 (2013).
De Mattos-Arruda, L. et al. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat. Rev. Clin. Oncol. 10, 377–389 (2013).
de Bono, J. S. et al. Circulating tumour cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14, 6302–6309 (2008).
Potapov, V. & Ong, J. L. Examining sources of error in PCR by single-molecule sequencing. PLoS ONE 12, e0181128 (2017).
Schirmer, M. et al. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43, e37 (2015).
Schirmer, M., Damore, R., Ijaz, U. Z., Hall, N. & Quince, C. Illumina error profiles: resolving fine-scale variation in metagenomic sequencing data. BMC Bioinformatics 17, 125 (2016).
Minoche, A. E., Dohm, J. C. & Himmelbauer, H. Evaluation of genomic high-throughput sequencing data generated on Illumina HiSeq and genome analyzer systems. Genome Biol. 12, R112 (2011).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Quail, M. A. et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341 (2012).
Leary, R. J. et al. Development of personalized tumour biomarkers using massively parallel sequencing. Sci. Transl. Med. 2, 20ra14 (2010).
Acknowledgements
We thank S. Skates for useful discussions regarding diagnostic economics and outcomes. L.N.K. was supported by NIH grant P01CA163222. A.A.P. was supported by NIH grants R01CA197486 and R01CA233364. D.Y.Z. was supported by NIH grants R01CA203964 and R01CA233364, and by CPRIT grant RP180147.
Author information
Authors and Affiliations
Contributions
P.S., L.R.W. and D.Y.Z. wrote the paper on the basis of in-depth discussions with all authors.
Corresponding author
Ethics declarations
Competing interests
P.S., L.R.W. and A.A.P. are consultants for NuProbe USA. A.A.P. is a consultant for Binary Genomics and has equity ownership in the company. D.Y.Z. is a co-founder of, and holds significant equity in, NuProbe Global, Torus Biosystems and Pana Bio.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Matlab scripts.
Rights and permissions
About this article
Cite this article
Song, P., Wu, L.R., Yan, Y.H. et al. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng 6, 232–245 (2022). https://doi.org/10.1038/s41551-021-00837-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00837-3
This article is cited by
-
Genomic spectrum of actionable alterations in serial cell free DNA (cfDNA) analysis of patients with metastatic breast cancer
npj Breast Cancer (2024)
-
Artificial urinary biomarker probes for diagnosis
Nature Reviews Bioengineering (2024)
-
Circulating Tumor DNA (ctDNA) and Its Role in Gynecologic Malignancies
Current Treatment Options in Oncology (2024)
-
Relationship between plasma circulating cell-free DNA concentration and treatment outcomes including prognosis in patients with advanced non-small cell lung cancer
BMC Pulmonary Medicine (2023)
-
The functional and clinical roles of liquid biopsy in patient-derived models
Journal of Hematology & Oncology (2023)