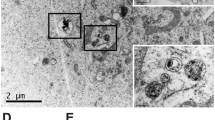
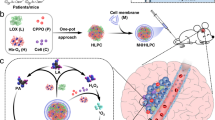
Abstract
In patients with glioblastoma, resistance to the chemotherapeutic temozolomide (TMZ) limits any survival benefits conferred by the drug. Here we show that the convection-enhanced delivery of nanoparticles containing disulfide bonds (which are cleaved in the reductive environment of the tumour) and encapsulating an oxaliplatin prodrug and a cationic DNA intercalator inhibit the growth of TMZ-resistant cells from patient-derived xenografts, and hinder the progression of TMZ-resistant human glioblastoma tumours in mice without causing any detectable toxicity. Genome-wide RNA profiling and metabolomic analyses of a glioma cell line treated with the cationic intercalator or with TMZ showed substantial differences in the signalling and metabolic pathways altered by each drug. Our findings suggest that the combination of anticancer drugs with distinct mechanisms of action with selective drug release and convection-enhanced delivery may represent a translational strategy for the treatment of TMZ-resistant gliomas.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The authors declare that the main data supporting the findings of this study are available within the paper and its Supplementary Information. The raw data generated for the RNA-seq analysis are available from the NCBI SRA database under the accession code PRJNA668337. The metabolomic dataset generated during the study is too large (2.3 GB) to be publicly shared, but the data are available for research purposes from the corresponding authors on reasonable request.
References
Stupp, R. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–466 (2009).
Dolecek, T. A., Propp, J. M., Stroup, N. E. & Kruchko, C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 14 (Suppl. 5), v1–49 (2012).
Kaina, B., Fritz, G., Mitra, S. & Coquerelle, T. Transfection and expression of human O6-methylguanine-DNA methyltransferase (MGMT) cDNA in Chinese hamster cells: the role of MGMT in protection against the genotoxic effects of alkylating agents. Carcinogenesis 12, 1857–1867 (1991).
Kitange, G. J. et al. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 11, 281–291 (2009).
Taylor, J. W. & Schiff, D. Treatment considerations for MGMT-unmethylated glioblastoma. Curr. Neurol. Neurosci. Rep. 15, 507 (2015).
Li, Q. J., Cai, J. Q. & Liu, C. Y. Evolving molecular genetics of glioblastoma. Chin. Med J. 129, 464–471 (2016).
Chen, X. et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat. Commun. 9, 2949 (2018).
Lapointe, S., Perry, A. & Butowski, N. A. Primary brain tumours in adults. Lancet 392, 432–446 (2018).
Xiao, H. H. et al. Recent progress in polymer-based platinum drug delivery systems. Prog. Polym. Sci. 87, 70–106 (2018).
Wang, S., Higgins, V. J., Aldrich-Wright, J. R. & Wu, M. J. Identification of the molecular mechanisms underlying the cytotoxic action of a potent platinum metallointercalator. J. Chem. Biol. 5, 51–61 (2012).
Graham, J., Mushin, M. & Kirkpatrick, P. Oxaliplatin. Nat. Rev. Drug Discov. 3, 11–12 (2004).
Pasetto, L. M., D’Andrea, M. R., Rossi, E. & Monfardini, S. Oxaliplatin-related neurotoxicity: how and why? Crit. Rev. Oncol. Hematol. 59, 159–168 (2006).
Golomb, L., Volarevic, S. & Oren, M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 588, 2571–2579 (2014).
Bruno, P. M. et al. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat. Med 23, 461–471 (2017).
Pisani, M. J., Wheate, N. J., Keene, F. R., Aldrich-Wright, J. R. & Collins, J. G. Anionic PAMAM dendrimers as drug delivery vehicles for transition metal-based anticancer drugs. J. Inorg. Biochem 103, 373–380 (2009).
Wheate, N. J. et al. Novel platinum(ii)-based anticancer complexes and molecular hosts as their drug delivery vehicles. Dalton Trans. 2007, 5055–5064 (2007).
Di Francia, R. et al. Current strategies to minimize toxicity of oxaliplatin: selection of pharmacogenomic panel tests. Anticancer Drugs 24, 1069–1078 (2013).
Jiang, Y. et al. SOD1 nanozyme with reduced toxicity and MPS accumulation. J. Control. Release 231, 38–49 (2016).
Jiang, Y., Brynskikh, A. M., Manickam, D. S. M. & Kabanov, A. V. SOD1 nanozyme salvages ischemic brain by locally protecting cerebral vasculature. J. Control. Release 213, 36–44 (2015).
Natarajan, G. et al. Nanoformulated copper/zinc superoxide dismutase exerts differential effects on glucose vs lipid homeostasis depending on the diet composition possibly via altered AMPK signaling. Transl. Res. 188, 10–26 (2017).
Caster, J. M., Patel, A. N., Zhang, T. & Wang, A. Investigational nanomedicines in 2016: a review of nanotherapeutics currently undergoing clinical trials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 9, 1416 (2017).
Liechty, W. B., Kryscio, D. R., Slaughter, B. V. & Peppas, N. A. Polymers for drug delivery systems. Annu Rev. Chem. Biomol. 1, 149–173 (2010).
Meng, F. H., Hennink, W. E. & Zhong, Z. Reduction-sensitive polymers and bioconjugates for biomedical applications. Biomaterials 30, 2180–2198 (2009).
Guo, X. et al. Advances in redox-responsive drug delivery systems of tumor microenvironment. J. Nanobiotechnology 16, 74 (2018).
Jiang, Y. et al. A ‘top-down’ approach to actuate poly(amine-co-ester) terpolymers for potent and safe mRNA delivery. Biomaterials 176, 122–130 (2018).
Kauffman, A. C. et al. Tunability of biodegradable poly(amine-co-ester) polymers for customized nucleic acid delivery and other biomedical applications. Biomacromolecules 19, 3861–3873 (2018).
Kuppusamy, P. et al. Noninvasive imaging of tumor redox status and its modification by tissue glutathione levels. Cancer Res. 62, 307–312 (2002).
Zhu, Z. et al. Glutathione reductase mediates drug resistance in glioblastoma cells by regulating redox homeostasis. J. Neurochem. 144, 93–104 (2018).
Efremenko, E. N. et al. A simple and highly effective catalytic nanozyme scavenger for organophosphorus neurotoxins. J. Control. Release 247, 175–181 (2017).
Harris, N. M. et al. Nano-particle delivery of brain derived neurotrophic factor after focal cerebral ischemia reduces tissue injury and enhances behavioral recovery. Pharm. Biochem. Behav. 150–151, 48–56 (2016).
Jiang, Y. et al. Nanoformulation of brain-derived neurotrophic factor with target receptor-triggered-release in the central nervous system. Adv. Funct. Mater. 28, 1703982 (2018).
Jahangiri, A. et al. Convection-enhanced delivery in glioblastoma: a review of preclinical and clinical studies. J. Neurosurg. 126, 191–200 (2017).
Bobo, R. H. et al. Convection-enhanced delivery of macromolecules in the brain. Proc. Natl Acad. Sci. USA 91, 2076–2080 (1994).
Fung, L. K. et al. Pharmacokinetics of interstitial delivery of carmustine, 4-hydroperoxycyclophosphamide, and paclitaxel from a biodegradable polymer implant in the monkey brain. Cancer Res 58, 672–684 (1998).
Kataoka, K., Harada, A. & Nagasaki, Y. Block copolymer micelles for drug delivery: design, characterization and biological significance. Adv. Drug Deliv. Rev. 47, 113–131 (2001).
Goodwin, A. P., Mynar, J. L., Ma, Y., Fleming, G. R. & Frechet, J. M. Synthetic micelle sensitive to IR light via a two-photon process. J. Am. Chem. Soc. 127, 9952–9953 (2005).
Su, H. et al. The role of critical micellization concentration in efficacy and toxicity of supramolecular polymers. Proc. Natl Acad. Sci. USA 117, 4518–4526 (2020).
Shang, L., Nienhaus, K. & Nienhaus, G. U. Engineered nanoparticles interacting with cells: size matters. J. Nanobiotechnology 12, 5 (2014).
Prabha, S., Arya, G., Chandra, R., Ahmed, B. & Nimesh, S. Effect of size on biological properties of nanoparticles employed in gene delivery. Artif. Cells Nanomed. Biotechnol. 44, 83–91 (2016).
He, C., Hu, Y., Yin, L., Tang, C. & Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 31, 3657–3666 (2010).
Mandl, H. K. et al. Optimizing biodegradable nanoparticle size for tissue-specific delivery. J. Control. Release 314, 92–101 (2019).
Wu, G., Fang, Y. Z., Yang, S., Lupton, J. R. & Turner, N. D. Glutathione metabolism and its implications for health. J. Nutr. 134, 489–492 (2004).
Griffith, O. W. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med 27, 922–935 (1999).
Meister, A. Glutathione metabolism and its selective modification. J. Biol. Chem. 263, 17205–17208 (1988).
Jones, D. P. et al. Glutathione measurement in human plasma. Evaluation of sample collection, storage and derivatization conditions for analysis of dansyl derivatives by HPLC. Clin. Chim. Acta 275, 175–184 (1998).
Snipstad, S. et al. Contact-mediated intracellular delivery of hydrophobic drugs from polymeric nanoparticles. Cancer Nanotechnol. 5, 8 (2014).
Korst, A. E., Boven, E., van der Sterre, M. L., Fichtinger-Schepman, A. M. & van der Vijgh, W. J. Influence of single and multiple doses of amifostine on the efficacy and the pharmacokinetics of carboplatin in mice. Br. J. Cancer 75, 1439–1446 (1997).
Paraskar, A., Soni, S., Roy, B., Papa, A. L. & Sengupta, S. Rationally designed oxaliplatin-nanoparticle for enhanced antitumor efficacy. Nanotechnology 23, 075103 (2012).
Haragsim, L. & Zima, T. Protective effects of verapamil on cis-platinum and carboplatinum nephrotoxicity in dehydrated and normohydrated rats. Biochem. Int. 28, 273–276 (1992).
Suttie, A. W. Histopathology of the spleen. Toxicol. Pathol. 34, 466–503 (2006).
Gupta, N., Lal, P., Vindal, A., Hadke, N. S. & Khurana, N. Spontaneous rupture of malarial spleen presenting as hemoperitoneum: a case report. J. Vector Borne Dis. 47, 119–120 (2010).
Toxicology and Carcinogenesis Studies of 3,3′,4,4′-Tetrachloroazobenzene (TCAB) (CAS No. 14047-09-7) in Harlan Sprague-Dawley Rats and B6C3F1 Mice (Gavage Studies) Techincal Report Series (National Toxicology Program, 2010).
Toxicology and Carcinogenesis Studies of α,β-Thujone (CAS No. 76231-76-0) in F344/N Rats and B6C3F1 Mice (Gavage Studies) Technical Report Series (National Toxicology Program, 2011).
Ward, J. M., Rehg, J. E. & Morse, H. C. 3rd Differentiation of rodent immune and hematopoietic system reactive lesions from neoplasias. Toxicol. Pathol. 40, 425–434 (2012).
Andreassen, P. R. & Ren, K. Fanconi anemia proteins, DNA interstrand crosslink repair pathways, and cancer therapy. Curr. Cancer Drug Targets 9, 101–117 (2009).
Thambi, T. et al. Bioreducible carboxymethyl dextran nanoparticles for tumor-targeted drug delivery. Adv. Health. Mater. 3, 1829–1838 (2014).
Son, S. et al. Anti-Trop2 antibody-conjugated bioreducible nanoparticles for targeted triple negative breast cancer therapy. Int J. Biol. Macromol. 110, 406–415 (2018).
Xia, W. et al. Bioreducible polyethylenimine-delivered siRNA targeting human telomerase reverse transcriptase inhibits HepG2 cell growth in vitro and in vivo. J. Control. Release 157, 427–436 (2012).
Florinas, S., Kim, J., Nam, K., Janat-Amsbury, M. M. & Kim, S. W. Ultrasound-assisted siRNA delivery via arginine-grafted bioreducible polymer and microbubbles targeting VEGF for ovarian cancer treatment. J. Control. Release 183, 1–8 (2014).
Lopez-Bertoni, H. et al. Bioreducible polymeric nanoparticles containing multiplexed cancer stem cell regulating miRNAs inhibit glioblastoma growth and prolong survival. Nano Lett. 18, 4086–4094 (2018).
Carlson, B. L., Pokorny, J. L., Schroeder, M. A. & Sarkaria, J. N. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr. Protoc. Pharmacol. https://doi.org/10.1002/0471141755.ph1416s52 (2011).
Vaubel, R. A. et al. Genomic and phenotypic characterization of a broad panel of patient-derived xenografts reflects the diversity of glioblastoma. Clin. Cancer Res. 26, 1094–1104 (2020).
Tew, B. Y. et al. Patient-derived xenografts of central nervous system metastasis reveal expansion of aggressive minor clones. Neuro Oncol. 22, 70–83 (2020).
Randall, E. C. et al. Localized metabolomic gradients in patient-derived xenograft models of glioblastoma. Cancer Res. 80, 1258–1267 (2020).
Randall, E. C. et al. Integrated mapping of pharmacokinetics and pharmacodynamics in a patient-derived xenograft model of glioblastoma. Nat. Commun. 9, 4904 (2018).
Schenone, M., Dancik, V., Wagner, B. K. & Clemons, P. A. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 9, 232–240 (2013).
Reichel, D. et al. Near infrared fluorescent nanoplatform for targeted intraoperative resection and chemotherapeutic treatment of glioblastoma. ACS Nano 14, 8392–8408 (2020).
Moser, V. C. Functional assays for neurotoxicity testing. Toxicol. Pathol. 39, 36–45 (2011).
Wu, T. et al. A nanobody-conjugated DNA nanoplatform for targeted platinum-drug delivery. Angew. Chem. Int. Ed. 58, 14224–14228 (2019).
Song, E. et al. Surface chemistry governs cellular tropism of nanoparticles in the brain. Nat. Commun. 8, 15322 (2017).
Serwer, L., Hashizume, R., Ozawa, T. & James, C. D. Systemic and local drug delivery for treating diseases of the central nervous system in rodent models. J. Vis. Exp. 42, 1992 (2010).
Xia, J. & Wishart, D. S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 6, 743–760 (2011).
Acknowledgements
This work was funded by grants from the US National Institutes of Health (CA149128 to W.M.S.), the National Natural Science Foundation of China (51873218 to H.X.), the National Science and Technology Major Project (2018ZX10734401 to H.X.), the Beijing Natural Science Foundation (2202071 to H.X.), and the Key Research and Development Program of Hunan Province (2019SK2251 to H.X.). A.S.P.-D. was supported by fellowships from NIH (T32 GM86287 and F32 HL142144) and the Cystic Fibrosis Foundation (PIOTRO20F0). A.J. was supported by a fellowship from the US National Science Foundation. We thank J. Ding for providing luciferase vectors.
Author information
Authors and Affiliations
Contributions
Y.W., R.S.B., H.X. and W.M.S. discussed and designed the study. D.W., Y.Y. and L.Z. prepared and characterized the polymer and drugs. P.S. and T.L. helped conduct the cell viability assays. H.K.M. assisted in analysing the flow cytometry data. A.S.P.-D. helped perform toxicity experiments. A.H. analysed the haematoxylin and eosin images from the brain. X.L. and Z.Z. aided in the metabolome analysis. A.J., Y.C., Y.Z., P.S. and F.W. contributed to characterization of NPs. X.C. and F.L. helped with the statistical analysis. Y.W. and Y.J. conducted all other experiments in this manuscript. All authors discussed the data and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks Shiv Gupta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures.
Supplementary Dataset 1
Gene information for Fig. 6a.
Supplementary Dataset 2
P values for comparisons conducted in Fig. 4.
Rights and permissions
About this article
Cite this article
Wang, Y., Jiang, Y., Wei, D. et al. Nanoparticle-mediated convection-enhanced delivery of a DNA intercalator to gliomas circumvents temozolomide resistance. Nat Biomed Eng 5, 1048–1058 (2021). https://doi.org/10.1038/s41551-021-00728-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00728-7
This article is cited by
-
Nanoparticles targeting mutant p53 overcome chemoresistance and tumor recurrence in non-small cell lung cancer
Nature Communications (2024)
-
Chemotactic nanomotor for multimodal combined therapy of glioblastoma
Science China Chemistry (2024)
-
Exosome-transmitted circCABIN1 promotes temozolomide resistance in glioblastoma via sustaining ErbB downstream signaling
Journal of Nanobiotechnology (2023)
-
METTL3 knockdown promotes temozolomide sensitivity of glioma stem cells via decreasing MGMT and APNG mRNA stability
Cell Death Discovery (2023)
-
The blood–brain barrier: structure, regulation, and drug delivery
Signal Transduction and Targeted Therapy (2023)