Abstract
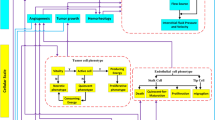
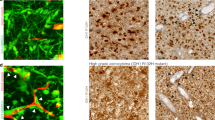
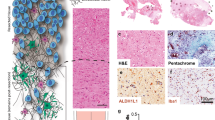
Glioblastoma stem-like cells dynamically transition between a chemoradiation-resistant state and a chemoradiation-sensitive state. However, physical barriers in the tumour microenvironment restrict the delivery of chemotherapy to tumour compartments that are distant from blood vessels. Here, we show that a massively parallel computational model of the spatiotemporal dynamics of the perivascular niche that incorporates glioblastoma stem-like cells and differentiated tumour cells as well as relevant tissue-level phenomena can be used to optimize the administration schedules of concurrent radiation and temozolomide—the standard-of-care treatment for glioblastoma. In mice with platelet-derived growth factor (PDGF)-driven glioblastoma, the model-optimized treatment schedule increased the survival of the animals. For standard radiation fractionation in patients, the model predicts that chemotherapy may be optimally administered about one hour before radiation treatment. Computational models of the spatiotemporal dynamics of the tumour microenvironment could be used to predict tumour responses to a broader range of treatments and to optimize treatment regimens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request.
Code availability
The custom code used in this study is available at GitHub (https://github.com/arandles/chemoradiation) under the BSD-3-Clause open-source license.
References
Weller, M. et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 18, 170–186 (2021).
Gramatzki, D. Glioblastoma in the Canton of Zurich, Switzerland revisited: 2005 to 2009. Cancer 122, 3740–3741 (2016).
Ostrom, Q. T. et al. CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro Oncol. 22, iv1–iv96 (2020).
Khan, L. et al. External beam radiation dose escalation for high grade glioma. Cochrane Database Syst. Rev. CD011475 https://doi.org/10.1002/14651858.CD011475.pub2 (2016).
Louis, D. N. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820 (2016).
Calabrese, C. et al. A perivascular niche for brain tumour stem cells. Cancer Cell 11, 69–82 (2007).
Eyler, C. E. et al. Glioma stem cell proliferation and tumour growth are promoted by nitric oxide synthase-2. Cell 146, 53–66 (2011).
Charles, N. et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell 6, 141–152 (2010).
Bleau, A.-M. et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumour stem-like cells. Cell Stem Cell 4, 226–235 (2009).
Kozin, S. V., Duda, D. G., Munn, L. L. & Jain, R. K. Neovascularization after irradiation: what is the source of newly formed vessels in recurring tumors? J. Natl Cancer Inst. 104, 899–905 (2012).
Garcia-Barros, M. et al. Tumor response to radiotherapy regulated by endothelial cell apoptosis. Science 300, 1155–1159 (2003).
Radbruch, A. et al. Quantification of tumour vessels in glioblastoma patients using time-of-flight angiography at 7 Tesla: a feasibility study. PLoS ONE 9, e110727 (2014).
Mustafa, D. et al. Expression sites of colligin 2 in glioma blood vessels. Brain Pathol. 20, 50–65 (2010).
Houghton, P. J. et al. Antitumor activity of temozolomide combined with irinotecan is partly independent of O6-methylguanine-DNA methyltransferase and mismatch repair phenotypes in xenograft models. Clin. Cancer Res. 6, 4110–4118 (2000).
Gilbert, M. R. et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J. Clin. Oncol. 31, 4085–4091 (2013).
Brada, M. et al. Temozolomide versus procarbazine, lomustine, and vincristine in recurrent high-grade glioma. J. Clin. Oncol. 28, 4601–4608 (2010).
Minchinton, A. I. & Tannock, I. F. Drug penetration in solid tumours. Nat. Rev. Cancer 6, 583–592 (2006).
Leder, K. Mathematical modelling of PDGF-driven glioblastoma reveals optimized radiation dosing schedules. Cell 7, 603–616 (2014).
Stevens, M. F. et al. Antitumor activity and pharmacokinetics in mice of 8-carbamoyl-3-methyl-imidazo[5,1-d]−1,2,3,5-tetrazin-4(3H)-one (CCRG 81045; M & B 39831), a novel drug with potential as an alternative to dacarbazine. Cancer Res. 47, 5846–5852 (1987).
Newlands, E. S. et al. Phase I trial of temozolomide (CCRG 81045: M&B 39831: NSC 362856). Br. J. Cancer 65, 287–291 (1992).
Ostermann, S. et al. Plasma and cerebrospinal fluid population pharmacokinetics of temozolomide in malignant glioma patients. Clin. Cancer Res. 10, 3728–3736 (2004).
Charles, N. A. & Holland, E. C. TRRAP and the maintenance of stemness in gliomas. Cell Stem Cell 6, 6–7 (2010).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Perry, J. R. et al. Short-course radiation plus temozolomide in elderly patients with glioblastoma. N. Engl. J. Med. 376, 1027–1037 (2017).
Agarwala, S. S. & Kirkwood, J. M. Temozolomide, a novel alkylating agent with activity in the central nervous system, may improve the treatment of advanced metastatic melanoma. Oncologist 5, 144–151 (2000).
Carlson, B. L. et al. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int. J. Radiat. Oncol. Biol. Phys. 75, 212–219 (2009).
Banissi, C., Ghiringhelli, F., Chen, L. & Carpentier, A. F. Treg depletion with a low-dose metronomic temozolomide regimen in a rat glioma model. Cancer Immunol. Immunother. 58, 1627–1634 (2009).
Weller, M. et al. MGMT promoter methylation is a strong prognostic biomarker for benefit from dose-intensified temozolomide rechallenge in progressive glioblastoma: the DIRECTOR trial. Clin. Cancer Res. 21, 2057–2064 (2015).
Thorsson, V. et al. The immune landscape of cancer. Immunity 48, 812–830 (2018).
Wang, J. et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 48, 768–776 (2016).
Snuderl, M. et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20, 810–817 (2011).
Körber, V. et al. Evolutionary trajectories of IDHWT glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell 35, 692–704 (2019).
Wang, Q. et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56 (2017).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Capper, D. et al. DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018).
Hu, X. et al. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma. Neoplasia 7, 356–368 (2005).
Mirams, G. R. et al. Chaste: an open source C++ library for computational physiology and biology. PLoS Comput. Biol. 9, e1002970 (2013).
Charles, N. & Holland, E. C. The perivascular niche microenvironment in brain tumour progression. Cell Cycle 9, 3012–3021 (2010).
Verlet, L. Computer ‘experiments’ on classical fluids. I. Thermodynamical properties of Lennard-Jones molecules. Phys. Rev. 159, 98–103 (1967).
Fuchs, E., Tumbar, T. & Guasch, G. Socializing with the neighbors: stem cells and their niche. Cell 116, 769–778 (2004).
Wedge, S. R., Porteous, J. K., Glaser, M. G., Marcus, K. & Newlands, E. S. In vitro evaluation of temozolomide combined with X-irradiation. Anticancer Drugs 8, 92–97 (1997).
Brock, C. S. et al. Phase I trial of temozolomide using an extended continuous oral schedule. Cancer Res. 58, 4363–4367 (1998).
Thomas, V., Kumari, T. V. & Jayabalan, M. In vitro studies on the effect of physical cross-linking on the biological performance of aliphatic poly(urethane urea) for blood contact applications. Biomacromolecules 2, 588–596 (2001).
Kirkpatrick, S., Gelatt, C. D. & Vecchi, M. P. Optimization by simulated annealing. Science 220, 671–680 (1983).
Aarts, E. & Korst, J. Simulated Annealing and Boltzmann Machines: A Stochastic Approach to Combinatorial Optimization and Neural Computing. (John Wiley & Sons, 1989).
Acknowledgements
We acknowledge feedback and advice from members of the Michor laboratory, and D. Puleri for figure editing. We also acknowledge support from the Dana-Farber Physical Sciences Oncology Center (NIH U54CA193461, to F.M. and E.C.H.), the NIH Office of the Director (NIH, DP5OD019876, to A.R.), and the Lawrence Livermore National Laboratory Lawrence Fellowship (to A.R.). The mouse work was supported by P30 CA015704 at the Fred Hutchinson Cancer Research Center. This work was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. Computing support for this work was provided by the Lawrence Livermore National Laboratory Institutional Computing Grand Challenges. F.M. acknowledges support from the Ludwig Center at Harvard.
Author information
Authors and Affiliations
Contributions
A.R., H.-G.W., E.C.H. and F.M. contributed to the design of the study. A.R., J.A.D. and Y.-K.C. performed the computational modelling. H.-G.W., S.E. and S.S.P. performed the mouse experiments. E.C.H. and F.M. supervised the study. All of the authors contributed to the writing of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures and tables.
Rights and permissions
About this article
Cite this article
Randles, A., Wirsching, HG., Dean, J.A. et al. Computational modelling of perivascular-niche dynamics for the optimization of treatment schedules for glioblastoma. Nat Biomed Eng 5, 346–359 (2021). https://doi.org/10.1038/s41551-021-00710-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00710-3
This article is cited by
-
Computational approaches to modelling and optimizing cancer treatment
Nature Reviews Bioengineering (2023)