Abstract
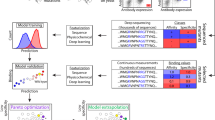
The optimization of therapeutic antibodies is time-intensive and resource-demanding, largely because of the low-throughput screening of full-length antibodies (approximately 1 × 103 variants) expressed in mammalian cells, which typically results in few optimized leads. Here we show that optimized antibody variants can be identified by predicting antigen specificity via deep learning from a massively diverse space of antibody sequences. To produce data for training deep neural networks, we deep-sequenced libraries of the therapeutic antibody trastuzumab (about 1 × 104 variants), expressed in a mammalian cell line through site-directed mutagenesis via CRISPR–Cas9-mediated homology-directed repair, and screened the libraries for specificity to human epidermal growth factor receptor 2 (HER2). We then used the trained neural networks to screen a computational library of approximately 1 × 108 trastuzumab variants and predict the HER2-specific subset (approximately 1 × 106 variants), which can then be filtered for viscosity, clearance, solubility and immunogenicity to generate thousands of highly optimized lead candidates. Recombinant expression and experimental testing of 30 randomly selected variants from the unfiltered library showed that all 30 retained specificity for HER2. Deep learning may facilitate antibody engineering and optimization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared; however, they are available for research purposes from the corresponding author on reasonable request.
Code availability
Deep-learning models were built in Python v3.6.5 using the Keras v2.1.6 Sequential model as a wrapper for TensorFlow v1.8.0. The code and models used to perform the work in this study are available at the following github repository: https://github.com/dahjan/DMS_opt.
References
Paul, S. M. et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 9, 203–214 (2010).
Sharma, V. K. et al. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc. Natl Acad. Sci. USA 111, 18601–18606 (2014).
Jain, T. et al. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl Acad. Sci. USA 114, 944–949 (2017).
Hu, D. et al. Effective optimization of antibody affinity by phage display integrated with high-throughput DNA synthesis and sequencing technologies. PLoS ONE 10, e0129125 (2015).
Bos, A. B. et al. Development of a semi-automated high throughput transient transfection system. J. Biotechnol. 180, 10–16 (2014).
Tomar, D. S., Kumar, S., Singh, S. K., Goswami, S. & Li, L. Molecular basis of high viscosity in concentrated antibody solutions: strategies for high concentration drug product development. mAbs 8, 216–228 (2016).
Roth, E. M. et al. Antidrug antibodies in patients treated with alirocumab. N. Engl. J. Med. 376, 1589–1590 (2017).
Greiff, V. et al. Learning the high-dimensional immunogenomic features that predict public and private antibody repertoires. J. Immunol. https://doi.org/10.4049/jimmunol.1700594 (2017).
Christensen, T., Frandsen, A., Glazier, S., Humpherys, J. & Kartchner, D. Machine learning methods for disease prediction with claims data. In 2018 IEEE International Conference on Healthcare Informatics https://doi.org/10.1109/ICHI.2018.00108 (IEEE, 2018).
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
Fox, R. et al. Optimizing the search algorithm for protein engineering by directed evolution. Protein Eng. Des. Sel. 16, 589–597 (2003).
Fox, R. Directed molecular evolution by machine learning and the influence of nonlinear interactions. J. Theor. Biol. 234, 187–199 (2005).
Romero, P. A., Krause, A. & Arnold, F. H. Navigating the protein fitness landscape with Gaussian processes. Proc. Natl Acad. Sci. USA 110, E193–E201 (2013).
Bedbrook, C. N., Yang, K. K., Rice, A. J., Gradinaru, V. & Arnold, F. H. Machine learning to design integral membrane channelrhodopsins for efficient eukaryotic expression and plasma membrane localization. PLoS Comput. Biol. 13, e1005786 (2017).
Angermueller, C., Pärnamaa, T., Parts, L. & Stegle, O. Deep learning for computational biology. Mol. Syst. Biol. 12, 878 (2016).
Wainberg, M., Merico, D., Delong, A. & Frey, B. J. Deep learning in biomedicine. Nat. Biotechnol. 36, 829–838 (2018).
Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015).
Cuperus, J. T. et al. Deep learning of the regulatory grammar of yeast 5′ untranslated regions from 500,000 random sequences. Genome Res. https://doi.org/10.1101/gr.224964.117 (2017).
Bulik-Sullivan, B. et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol. 37, 55–63 (2019).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Rosenblatt, F. The Perceptron, a Perceiving and Recognizing Automation Report 85-60-1 (Cornell Aeronautical Laboratory, 1957).
Pogson, M., Parola, C., Kelton, W. J., Heuberger, P. & Reddy, S. T. Immunogenomic engineering of a plug-and-(dis)play hybridoma platform. Nat. Commun. 7, 12535 (2016).
Mason, D. M. et al. High-throughput antibody engineering in mammalian cells by CRISPR/Cas9-mediated homology-directed mutagenesis. Nucleic Acids Res. https://doi.org/10.1093/nar/gky550 (2018).
Whitehead, T. A. et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 30, 543–548 (2012).
Cho, H.-S. et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 421, 756–760 (2003).
Rose, A. S. et al. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics 34, 3755–3758 (2018).
Miho, E., Roškar, R., Greiff, V. & Reddy, S. T. Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat. Commun. 10, 1321 (2019).
Sundararajan, M., Taly, A. & Yan, Q. Axiomatic attribution for deep networks. Preprint at https://arxiv.org/abs/1703.01365 (2017).
Sormanni, P., Aprile, F. A. & Vendruscolo, M. The CamSol method of rational design of protein mutants with enhanced solubility. J. Mol. Biol. 427, 478–490 (2015).
Pérez, A.-M. W. et al. In vitro and in silico assessment of the developability of a designed monoclonal antibody library. mAbs 11, 388–400 (2019).
Jensen, K. K. et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406 (2018).
Greenbaum, J. et al. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 63, 325–335 (2011).
Wu, Z., Kan, S. B. J., Lewis, R. D., Wittmann, B. J. & Arnold, F. H. Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1901979116 (2019).
Vajdos, F. F. et al. Comprehensive functional maps of the antigen-binding site of an anti-ErbB2 antibody obtained with shotgun scanning mutagenesis. J. Mol. Biol. 320, 415–428 (2002).
Townsend, S. et al. Augmented binary substitution: single-pass CDR germ-lining and stabilization of therapeutic antibodies. Proc. Natl Acad. Sci. USA 112, 15354–15359 (2015).
Trudeau, D. L., Smith, M. A. & Arnold, F. H. Innovation by homologous recombination. Curr. Opin. Chem. Biol. 17, 902–909 (2013).
Riesselman, A. J., Ingraham, J. B. & Marks, D. S. Deep generative models of genetic variation capture the effects of mutations. Nat. Methods 15, 816–822 (2018).
Sormanni, P., Aprile, F. A. & Vendruscolo, M. Third generation antibody discovery methods: in silico rational design. Chem. Soc. Rev. 47, 9137–9157 (2018).
Raybould, M. I. J. et al. Five computational developability guidelines for therapeutic antibody profiling. Proc. Natl Acad. Sci. USA 116, 4025–4030 (2019).
Rabia, L. A., Zhang, Y., Ludwig, S. D., Julian, M. C. & Tessier, P. M. Net charge of antibody complementarity-determining regions is a key predictor of specificity. Protein Eng. Des. Sel. https://doi.org/10.1093/protein/gzz002 (2019)
Abhinandan, K. R. & Martin, A. C. R. Analyzing the “degree of humanness” of antibody sequences. J. Mol. Biol. 369, 852–862 (2007).
van Brummelen, E. M. J., Ros, W., Wolbink, G., Beijnen, J. H. & Schellens, J. H. M. Antidrug antibody formation in oncology: clinical relevance and challenges. Oncologist 21, 1260–1268 (2016).
Vaisman-Mentesh, A., Gutierrez-Gonzalez, M., DeKosky, B. J. & Wine, Y. The molecular mechanisms that underlie the immune biology of anti-drug antibody formation following treatment with monoclonal antibodies. Front. Immunol. 11, 1951 (2020).
Igawa, T. et al. Antibody recycling by engineered pH-dependent antigen binding improves the duration of antigen neutralization. Nat. Biotechnol. 28, 1203–1207 (2010).
Kang, J. C. et al. Engineering a HER2-specific antibody–drug conjugate to increase lysosomal delivery and therapeutic efficacy. Nat. Biotechnol. https://doi.org/10.1038/s41587-019-0073-7 (2019).
Slaga, D. et al. Avidity-based binding to HER2 results in selective killing of HER2-overexpressing cells by anti-HER2/CD3. Sci. Transl. Med. 10, eaat5775 (2018).
Gainza, P. et al. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 17, 184–192 (2020).
Menzel, U. et al. Comprehensive evaluation and optimization of amplicon library preparation methods for high-throughput antibody sequencing. PLoS ONE 9, e96727 (2014).
Bolotin, D. A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015).
R Core Development Team. R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2014).
van Rossum, G. & Drake, F. L. The Python Language Reference Manual (Network Theory Ltd., 2011).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer International Publishing, 2016).
Brewer, C. A., Hatchard, G. W. & Harrower, M. A. ColorBrewer in print: a catalog of color schemes for maps. Cartogr. Geogr. Inf. Sci. 30, 5–32 (2003).
Wagih, O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647 (2017).
Fowler, D. M. et al. High-resolution mapping of protein sequence-function relationships. Nat. Methods 7, 741–746 (2010).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Machine Learning Res. 12, 2825–2830 (2011).
Chollet, F. Keras. https://keras.io (2015).
Abadi, M. et al. TensorFlow: a system for large-scale machine learning. In Proceedings of the 12th USENIX Conference on Operating Systems Design and Implementation 265–283 (USENIX Association, 2016).
Sokolova, M. & Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 45, 427–437 (2009).
Boughorbel, S., Jarray, F. & El-Anbari, M. Optimal classifier for imbalanced data using Matthews Correlation Coefficient metric. PLoS ONE 12, e0177678 (2017).
Csárdi, G. & Nepusz, T. The igraph software package for complex network research. InterJournal 1695 (2006).
Tareen, A. & Kinney, J. B. Logomaker: beautiful sequence logos in Python. Bioinformatics 36, 2272–2274 (2020).
Lide, D. R. Handbook of Chemistry and Physics 72nd edn (CRC Press, 1991).
Acknowledgements
We thank the ETH Zurich D-BSSE Single Cell Unit and the ETH Zurich D-BSSE Genomics Facility for support—in particular M. Di Tacchio, A. Gumienny, E. Burcklen and C. Beisel. We also thank the Vendruscolo Laboratory (Cambridge, UK), P. Sormanni in particular, for assistance with implementing the CamSol method on large libraries as well as the group of M. Nielson (DTU, Denmark) for providing an easy-to-use package for MHC class II affinity predictions. Funding was provided by the National Competence Center for Research on Molecular Systems Engineering.
Author information
Authors and Affiliations
Contributions
D.M.M., S.F., C.R.W. and S.T.R. developed the methodology. D.M.M. and S.T.R. designed the experiments and wrote the manuscript. D.M.M., C.R.W., S.F. and J.D. analysed the sequencing data and performed deep-learning analyses. P.G. and B.E.C. designed and performed the structural modelling experiments and analysis. C.J. generated in silico libraries. D.M.M. and R.A.E. performed experiments. B.W., S.M.M. and L.B. performed the cell-line development.
Corresponding author
Ethics declarations
Competing interests
ETH Zurich has filed for patent protection on the technology described herein, and D.M.M., S.F., C.R.W. and S.T.R. are named as co-inventors on this patent (International Filing Application PCT/IB2020/053370).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, figures and tables.
Rights and permissions
About this article
Cite this article
Mason, D.M., Friedensohn, S., Weber, C.R. et al. Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning. Nat Biomed Eng 5, 600–612 (2021). https://doi.org/10.1038/s41551-021-00699-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-021-00699-9
This article is cited by
-
Machine learning for functional protein design
Nature Biotechnology (2024)
-
Adaptive immune receptor repertoire analysis
Nature Reviews Methods Primers (2024)
-
Efficient evolution of human antibodies from general protein language models
Nature Biotechnology (2024)
-
Revolutionizing Synthetic Antibody Design: Harnessing Artificial Intelligence and Deep Sequencing Big Data for Unprecedented Advances
Molecular Biotechnology (2024)
-
Machine learning optimization of candidate antibody yields highly diverse sub-nanomolar affinity antibody libraries
Nature Communications (2023)