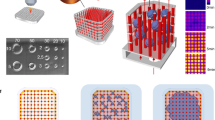
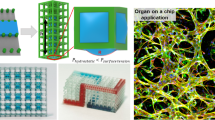
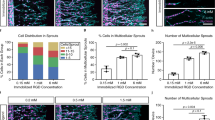
Abstract
Sacrificial templates for patterning perfusable vascular networks in engineered tissues have been constrained in architectural complexity, owing to the limitations of extrusion-based 3D printing techniques. Here, we show that cell-laden hydrogels can be patterned with algorithmically generated dendritic vessel networks and other complex hierarchical networks by using sacrificial templates made from laser-sintered carbohydrate powders. We quantified and modulated gradients of cell proliferation and cell metabolism emerging in response to fluid convection through these networks and to diffusion of oxygen and metabolites out of them. We also show scalable strategies for the fabrication, perfusion culture and volumetric analysis of large tissue-like constructs with complex and heterogeneous internal vascular architectures. Perfusable dendritic networks in cell-laden hydrogels may help sustain thick and densely cellularized engineered tissues, and assist interrogations of the interplay between mass transport and tissue function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Much of the source and analysed data are available in Zenodo (https://doi.org/10.5281/zenodo.3723373). Some source datasets are too large to be shared in public repositories and are available from the corresponding author on reasonable request. Design files and documentation for our open-source selective laser sintering hardware and software are available in the Zenodo repository and at https://github.com/MillerLabFTW/OpenSLS.
Code availability
A custom Python add-on for Blender to generate bifurcating vascular structures is available in the Zenodo repository and at https://github.com/MillerLabFTW/IntussusceptionAddon. Image-processing and analysis scripts are also available in the Zenodo repository. The mutual tree attraction algorithm for generating dendritic networks is closed source, but the generated architectures are included in the Zenodo repository.
Change history
16 June 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41551-021-00761-6
References
Zamir, M. Fractal dimensions and multifractility in vascular branching. J. Theor. Biol. 212, 183–190 (2001).
West, G. B., Brown, J. H. & Enquist, B. J. A general model for the origin of allometric scaling laws in biology. Science 276, 122–126 (1997).
West, G. B., Brown, J. H. & Enquist, B. J. A general model for ontogenetic growth. Nature 413, 628–631 (2001).
Monahan-Earley, R., Dvorak, A. M. & Aird, W. C. Evolutionary origins of the blood vascular system and endothelium. J. Thromb. Haemost. 11, 46–66 (2013).
Novosel, E. C., Kleinhans, C. & Kluger, P. J. Vascularization is the key challenge in tissue engineering. Adv. Drug Deliv. Rev. 63, 300–311 (2011).
Kinstlinger, I. S. & Miller, J. S. 3D-printed fluidic networks as vasculature for engineered tissue. Lab Chip 16, 2025–2043 (2016).
Cabodi, M. et al. A microfluidic biomaterial. J. Am. Chem. Soc. 127, 13788–13789 (2005).
Chrobak, K. M., Potter, D. R. & Tien, J. Formation of perfused, functional microvascular tubes in vitro. Microvasc. Res. 71, 185–196 (2006).
Zhang, B. et al. Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat. Mater. 15, 669–678 (2016).
Miller, J. S. The billion cell construct: will three-dimensional printing get us there? PLoS Biol. 12, e1001882 (2014).
Luo, Y., Lode, A. & Gelinsky, M. Direct plotting of three-dimensional hollow fiber scaffolds based on concentrated alginate pastes for tissue engineering. Adv. Healthc. Mater. 2, 777–783 (2013).
Christensen, K. et al. Freeform inkjet printing of cellular structures with bifurcations. Biotechnol. Bioeng. 112, 1047–1055 (2015).
Hinton, T. J. et al. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 1, e1500758 (2015).
Lee, A. et al. 3D bioprinting of collagen to rebuild components of the human heart. Science 365, 482–487 (2019).
Zhang, R. & Larsen, N. B. Stereolithographic hydrogel printing of 3D culture chips with biofunctionalized complex 3D perfusion networks. Lab Chip 17, 4273–4282 (2017).
Meyer, W. et al. Soft polymers for building up small and smallest blood supplying systems by stereolithography. J. Funct. Biomater. 3, 257–268 (2012).
Brandenberg, N. & Lutolf, M. P. In situ patterning of microfluidic networks in 3D cell-laden hydrogels. Adv. Mater. 28, 7450–7456 (2016).
Heintz, K. A. et al. Fabrication of 3D biomimetic microfluidic networks in hydrogels. Adv. Healthc. Mater. 5, 2153–2160 (2016).
Arakawa, C. K., Badeau, B. A., Zheng, Y. & DeForest, C. A. Multicellular vascularized engineered tissues through user-programmable biomaterial photodegradation. Adv. Mat. 29, 1703156 (2017).
Grigoryan, B. et al. Functional intravascular topologies and multivascular networks within biocompatible hydrogels. Science 364, 458–464 (2019).
Golden, A. P. & Tien, J. Fabrication of microfluidic hydrogels using molded gelatin as a sacrificial element. Lab Chip 7, 720–725 (2007).
Miller, J. S. et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 11, 768–774 (2012).
Bégin-Drolet, A. et al. Design of a 3D printer head for additive manufacturing of sugar glass for tissue engineering applications. Addit. Manuf. 15, 29–39 (2017).
Gelber, M. K., Hurst, G., Comi, T. J. & Bhargava, R. Model-guided design and characterization of a high-precision 3D printing process for carbohydrate glass. Addit. Manuf. 22, 38–50 (2018).
Homan, K. A. et al. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep. 6, 34845 (2016).
Kolesky, D. B. et al. 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv. Mater. 26, 3124–3130 (2014).
Kolesky, D. B., Homan, K. A., Skylar-Scott, M. A. & Lewis, J. A. Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl Acad. Sci. USA 113, 3179–3184 (2016).
Skylar-Scott, M. A. et al. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 5, eaaw2459 (2019).
Wu, W., Deconinck, A. & Lewis, J. A. Omnidirectional printing of 3D microvascular networks. Adv. Mater. 23, H178–H183 (2011).
Song, K. H., Highley, C. B., Rouff, A. & Burdick, J. A. Complex 3D-printed microchannels within cell-degradable hydrogels. Adv. Funct. Mater. 28, 1801331 (2018).
Pimentel, C. R. et al. Three-dimensional fabrication of thick and densely populated soft constructs with complex and actively perfused channel network. Acta Biomater. 65, 174–184 (2018).
Kinstlinger, I. S. et al. Open-source selective laser sintering (OpenSLS) of nylon and biocompatible polycaprolactone. PLoS ONE 11, e0147399 (2016).
Roszelle, B. N. et al. Flow diverter effect on cerebral aneurysm hemodynamics: An in vitro comparison of telescoping stents and the Pipeline. Neuroradiology 55, 751–758 (2013).
Saggiomo, V. & Velders, A. H. Simple 3D printed scaffold-removal method for the fabrication of intricate microfluidic devices. Adv. Sci. 2, 1500125 (2015).
Nguyen, L. H. et al. Vascularized bone tissue engineering: approaches for potential improvement. Tissue Eng. Part B 18, 363–382 (2012).
Nguyen, Q. T., Hwang, Y., Chen, A. C., Varghese, S. & Sah, R. L. Cartilage-like mechanical properties of poly (ethylene glycol)-diacrylate hydrogels. Biomaterials 33, 6682–6690 (2012).
Partlow, B. P. et al. Highly tunable elastomeric silk biomaterials. Adv. Funct. Mater. 24, 4615–4624 (2014).
Mooney, R., Tawil, B. & Mahoney, M. Specific fibrinogen and thrombin concentrations promote neuronal rather than glial growth when primary neural cells are seeded within plasma-derived fibrin gels. Tissue Eng. Part A 16, 1607–1619 (2010).
Duong, H., Wu, B. & Tawil, B. Modulation of 3D fibrin matrix stiffness by intrinsic fibrinogen–thrombin compositions and by extrinsic cellular activity. Tissue Eng. Part A 15, 1865–1876 (2009).
Subhash, G., Liu, Q., Moore, D. F., Ifju, P. G. & Haile, M. A. Concentration dependence of tensile behavior in agarose gel using digital image correlation. Exp. Mech. 51, 255–262 (2011).
Feugier, F. G., Mochizuki, A. & Iwasa, Y. Self-organization of the vascular system in plant leaves: inter-dependent dynamics of auxin flux and carrier proteins. J. Theor. Biol. 236, 366–375 (2005).
Fujita, H. & Mochizuki, A. The origin of the diversity of leaf venation pattern. Dev. Dyn. 235, 2710–2721 (2006).
Runions, A., Lane, B. & Prusinkiewicz, P. Modeling trees with a space colonization algorithm. In Proc. 3rd Eurographics Conference on Natural Phenomena (Eds Ebert, D. & Mérillou, S.) 63–70 (Eurographics Association, 2007).
Murray, C. D. The physiological principle of minimum work applied to the angle of branching of arteries. J. Gen. Physiol. 9, 835–841 (1926).
Miguel, A. F. Dendritic design as an archetype for growth patterns in nature: fractal and constructal views. Front. Phys. 2, 9 (2014).
Moon, J. J. et al. Biomimetic hydrogels with pro-angiogenic properties. Biomaterials 31, 3840–3847 (2010).
Calderon, G. et al. Tubulogenesis of co-cultured human iPS-derived endothelial cells and human mesenchymal stem cells in fibrin and gelatin methacrylate gels. Biomater. Sci. 5, 1652–1660 (2017).
Eskin, S. G., Ives, C., McIntire, L. & Navarro, L. Response of cultured endothelial cells to steady flow. Microvasc. Res. 28, 87–94 (1984).
Yang, P. J. & Temenoff, J. S. Engineering orthopedic tissue interfaces. Tissue Eng. Part B 15, 127–141 (2009).
Eckes, B. et al. Fibroblast-matrix interactions in wound healing and fibrosis. Matrix Biol. 19, 325–332 (2000).
Lu, P., Weaver, V. M. & Werb, Z. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196, 395–406 (2012).
Radisic, M. et al. Oxygen gradients correlate with cell density and cell viability in engineered cardiac tissue. Biotechnol. Bioeng. 93, 332–343 (2006).
Tocchio, A. et al. Versatile fabrication of vascularizable scaffolds for large tissue engineering in bioreactor. Biomaterials 45, 124–131 (2015).
Tsang, V. L. et al. Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels. FASEB J. 21, 790–801 (2007).
Krogh, A. The number and distribution of capillaries in muscles with calculations of the oxygen pressure head necessary for supplying the tissue. J. Physiol. 52, 409–415 (1919).
Lewis, M. C., MacArthur, B. D., Malda, J., Pettet, G. & Please, C. P. Heterogeneous proliferation within engineered cartilaginous tissue: the role of oxygen tension. Biotechnol. Bioeng. 91, 607–615 (2005).
Demol, J., Lambrechts, D., Geris, L., Schrooten, J. & Van Oosterwyck, H. Towards a quantitative understanding of oxygen tension and cell density evolution in fibrin hydrogels. Biomaterials 32, 107–118 (2011).
Gu, W. Y., Yao, H., Huang, C. Y. & Cheung, H. S. New insight into deformation-dependent hydraulic permeability of gels and cartilage, and dynamic behavior of agarose gels in confined compression. J. Biomech. 36, 593–598 (2003).
Chuppa, S. et al. Fermentor temperature as a tool for control of high-density perfusion cultures of mammalian cells. Biotechnol. Bioeng. 55, 328–338 (1997).
Ducommun, P., Ruffieux, P. A., Kadouri, A., Von Stockar, U. & Marison, I. W. Monitoring of temperature effects on animal cell metabolism in a packed bed process. Biotechnol. Bioeng. 77, 838–842 (2002).
Jorjani, P. & Ozturk, S. S. Effects of cell density and temperature on oxygen consumption rate for different mammalian cell lines. Biotechnol. Bioeng. 64, 349–356 (1999).
Xiang, C. et al. Long-term functional maintenance of primary human hepatocytes in vitro. Science 364, 399–402 (2019).
Bhatia, S. N., Balis, U. J., Yarmush, M. L. & Toner, M. Effect of cell–cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 13, 1883–1900 (1999).
Stevens, K. R. et al. InVERT molding for scalable control of tissue microarchitecture. Nat. Commun. 4, 1847 (2013).
Stevens, K. R. et al. In situ expansion of engineered human liver tissue in a mouse model of chronic liver disease. Sci. Transl. Med. 9, eaah5505 (2017).
Khetani, S. R. & Bhatia, S. N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 26, 120–126 (2008).
Bhatia, S. N., Underhill, G. H., Zaret, K. S. & Fox, I. J. Cell and tissue engineering for liver disease. Sci. Transl. Med. 6, 245sr2 (2014).
Rafii, S., Butler, J. M. & Ding, B.-S. Angiocrine functions of organ-specific endothelial cells. Nature 529, 316–325 (2016).
Baranski, J. D. et al. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc. Natl Acad. Sci USA 110, 7586–7591 (2013).
Mirabella, T. et al. 3D-printed vascular networks direct therapeutic angiogenesis in ischaemia. Nat. Biomed. Eng. 1, 0083 (2017).
Lindström, N. O. et al. Conserved and divergent molecular and anatomic features of human and mouse nephron patterning. J. Am. Soc. Nephrol. 29, 825–840 (2018).
Bosco, D. et al. Unique arrangement of α- and β-cells in human islets of Langerhans. 59, 1202–1210 (2010).
Wang, X.-N. et al. A three-dimensional atlas of human dermal leukocytes, lymphatics, and blood vessels. J. Invest. Dermatol. 134, 965–974 (2014).
Kang, H.-W. et al. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 34, 312–319 (2016).
Shapiro, A. J. et al. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regime. N. Engl. J. Med. 343, 230–238 (2000).
Iansante, V., Mitry, R. R., Filippi, C., Fitzpatrick, E. & Dhawan, A. Human hepatocyte transplantation for liver disease: current status and future perspectives. Pediatr. Res. 83, 232–240 (2018).
Parker Ponder, K. et al. Mouse hepatocytes migrate to liver parenchyma and function indefinitely after intrasplenic transplantation. Proc. Natl Acad. Sci. USA 88, 1217–1221 (1991).
Truslow, J. G. & Tien, J. Perfusion systems that minimize vascular volume fraction in engineered tissues. Biomicrofluidics 5, 022201 (2011).
Ronellenfitsch, H. & Katifori, E. Global optimization, local adaptation, and the role of growth in distribution networks. Phys. Rev. Lett. 117, 138301 (2016).
Freeman, R. Measuring the flow properties of consolidated, conditioned and aerated powders—a comparative study using a powder rheometer and a rotational shear cell. Powder Technol. 174, 25–33 (2007).
Thadavirul, N., Pavasant, P. & Supaphol, P. Development of polycaprolactone porous scaffolds by combining solvent casting, particulate leaching, and polymer leaching techniques for bone tissue engineering. J. Biomed. Mater. Res. A 102, 3379–3392 (2013).
Miller, J. S. et al. Bioactive hydrogels made from step-growth derived PEG-peptide macromers. Biomaterials 31, 3736–3743 (2010).
Rockwood, D. N. et al. Materials fabrication from Bombyx mori silk fibroin. Nat. Protoc. 6, 1612–1631 (2011).
Li, W., Germain, R. N. & Gerner, M. Y. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D).Proc. Natl Acad. Sci. USA 114, E7321–E7330 (2017).
Thielicke, W. & Stamhuis, E. J. PIVlab—towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw. 2, e30 (2014).
Thielicke, W. The Flapping Flight of Birds—Analysis and Application. PhD thesis, Rijksuniversiteit Groningen (2014).
Thielicke, W. & Stamhuis, E. J. PIVlab—time-resolved digital particle image velocimetry tool for MATLAB https://doi.org/10.6084/M9.FIGSHARE.1092508.V5 (2014).
Cheng, N.-S. Formula for the viscosity of a glycerol–water mixture. Ind. Eng. Chem. Res. 47, 3285–3288 (2008).
Volk, A. & Kähler, C. J. Density model for aqueous glycerol solutions. Exp. Fluids 59, 75 (2018).
van de Loosdrecht, A. A., Beelen, R. H., Ossenkoppele, G. J., Broekhoven, M. G. & Langenhuijsen, M. M. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J. Immunol. Methods 174, 311–320 (1994).
Ahrens, J., Geveci, B. & Law, C. in The Visualization Handbook (eds Hansen, C. D. & Johnson, C. R.) 717–731 (2005).
Acknowledgements
We thank A. Bastian, A. Ta and T. Schmidt for assistance with OpenSLS hardware and firmware; E. Watson and A. Mikos for assistance with mechanical testing; J. Wagner, P. Desai and C. F. Higgs for assistance with powder rheology; D. De Santos for technical assistance with carbohydrate SLS; D. Kaplan and W. Stoppel for providing silk fibroin; D. L. Gibbons for providing the 344SQ lung adenocarcinoma cell line; and C. Fortin for help with hepatocyte isolations. This work was supported in part by a Medical Research Grant from the Robert J. Kleberg Jr and Helen C. Kleberg Foundation (J.S.M.), National Institutes of Health (grants HL134510 and DK115461 (K.-D.B.)), the Texas Hepatocellular Carcinoma Consortium (THCCC) (CPRIT RP150587 (K.-D.B.)), National Insitutes of Health grant DP2HL137188 (K.R.S.) and National Insitutes of Health NIBIB Cardiovascular Training grant (T32EB001650 (S.H.S.)). I.S.K. acknowledges support by an F31 National Research Service Award (NRSA) from the National Institutes of Health (HL140905). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
I.S.K. and J.S.M. conceived and initiated the project and wrote the manuscript. I.S.K., S.H.S., G.A.C., K.V.R., D.R.Y., P.R.D., K.D.J. and F.J. designed and performed experiments. I.S.K., S.H.S. and G.A.C. acquired and analysed imaging data. J.E.R., J.D.L.-R. and S.S.P. developed generative design algorithms. D.W.S. synthesized materials. K.-D.B., K.R.S. and J.S.M. supervised the project.
Corresponding author
Ethics declarations
Competing interests
J.S.M. is a co-founder and holds an equity stake in Volumetric, Inc. J.E.R. and J.D.L.-R. are co-founders and hold equity stakes in Nervous System, Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods.
Supplementary Video 1
Annotated video recording of one layer of carbohydrate SLS.
Supplementary Video 2
Mutual-tree-attraction algorithm for the generative design of dendritic networks.
Supplementary Video 3
Fluorescent bead perfusion through a whole planar dendritic network.
Supplementary Video 4
Magnified view of fluorescent bead perfusion through the centre of a planar dendritic network.
Supplementary Video 5
Perfusion of dendritic architectures at a high volumetric flow rate.
Supplementary Video 6
Animated volume rendering of a region in an endothelialized planar dendritic network.
Supplementary Video 7
Rotating rendering of a volumetric μCT scan of a dendritic carbohydrate template.
Supplementary Video 8
Fly-through rendering of a volumetric μCT scan of a dendritic carbohydrate template.
Supplementary Video 9
Fly-through sequence of MTT staining in sections from a cell-laden gel with dendritic architecture.
Supplementary Video 10
Fly-through sequence of nuclear staining in sections from a cell-laden gel with dendritic architecture.
Supplementary Video 11
Fly-through sequence of processed MTT staining images from a cell-laden gel with dendritic architecture.
Supplementary Video 12
Rotating rendering of a volumetrically reconstructed MTT signal in a cell-laden gel with dendritic architecture.
Supplementary Video 13
Computational fluid-dynamics simulation of perfusion through a 3D dendritic architecture.
Rights and permissions
About this article
Cite this article
Kinstlinger, I.S., Saxton, S.H., Calderon, G.A. et al. Generation of model tissues with dendritic vascular networks via sacrificial laser-sintered carbohydrate templates. Nat Biomed Eng 4, 916–932 (2020). https://doi.org/10.1038/s41551-020-0566-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-020-0566-1
This article is cited by
-
Animated ink and wash character modeling and simulation based on automatic intelligent matching of local pixel blocks
Neural Computing and Applications (2023)
-
Engineering the next generation of cell-based therapeutics
Nature Reviews Drug Discovery (2022)
-
Self-shrinking soft demoulding for complex high-aspect-ratio microchannels
Nature Communications (2022)
-
Engineering the multiscale complexity of vascular networks
Nature Reviews Materials (2022)
-
Perfusion and endothelialization of engineered tissues with patterned vascular networks
Nature Protocols (2021)