Abstract
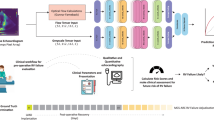
Machine learning promises to assist physicians with predictions of mortality and of other future clinical events by learning complex patterns from historical data, such as longitudinal electronic health records. Here we show that a convolutional neural network trained on raw pixel data in 812,278 echocardiographic videos from 34,362 individuals provides superior predictions of one-year all-cause mortality. The model’s predictions outperformed the widely used pooled cohort equations, the Seattle Heart Failure score (measured in an independent dataset of 2,404 patients with heart failure who underwent 3,384 echocardiograms), and a machine learning model involving 58 human-derived variables from echocardiograms and 100 clinical variables derived from electronic health records. We also show that cardiologists assisted by the model substantially improved the sensitivity of their predictions of one-year all-cause mortality by 13% while maintaining prediction specificity. Large unstructured datasets may enable deep learning to improve a wide range of clinical prediction models.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. All requests for raw and analysed data will be reviewed by the Legal Department of Geisinger Clinic to verify whether the request is subject to any intellectual property or confidentiality constraints. Requests for patient-related data not included in the paper will not be considered. Any data that can be shared will be released via a Material Transfer Agreement for non-commercial research purposes.
Code availability
All requests for code will be reviewed by the Legal Department of Geisinger Clinic to verify whether the request is subject to any intellectual property or confidentiality constraints. Any code that can be shared will be released via a Material Transfer Agreement for non-commercial research purposes under the Creative Commons Attribution NonCommercial–NoDerivatives 4.0 license. Code to reproduce Supplementary Fig. 10 is available at: https://github.com/alvarouc/geisinger-echo-mortality.
References
Payne, J. W. Task complexity and contingent processing in decision making: an information search and protocol analysis. Organ. Behav. Hum. Perform. 16, 366–387 (1976).
Quer, G., Muse, E. D., Nikzad, N., Topol, E. J. & Steinhubl, S. R. Augmenting diagnostic vision with AI. Lancet 390, 221 (2017).
Jha, S. & Topol, E. J. Adapting to artificial intelligence: radiologists and pathologists as information specialists. JAMA 316, 2353–2354 (2016).
Kyriacou, E., Constantinides, A., Pattichis, C., Pattichis, M. & Panayides, A. in Biomedical Signals, Imaging, and Informatics 4th edn (eds Bronzino, J. D. & Peterson, D.) Ch. 64 (CRC Press, 2015).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Ji, S., Xu, W., Yang, M. & Yu, K. 3D convolutional neural networks for human action recognition. IEEE Trans. Pattern Anal. Mach. Intell. 35, 221–231 (2012).
Karpathy, A. et al. Large-scale video classification with convolutional neural networks. In IEEE conference on Computer Vision and Pattern Recognition 1725–1732 (2014).
Gulshan, V. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Setio, A. A. A. et al. Pulmonary nodule detection in ct images: false positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging 35, 1160–1169 (2016).
Arbabshirani, M. R. et al. Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. npj Digit. Med. 1, 9 (2018).
Dou, Q. et al. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans. Med. Imaging 35, 1182–1195 (2016).
Madani, A., Ong, J. R., Tibrewal, A. & Mofrad, M. R. Deep echocardiography: data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. npj Digit. Med. 1, 59 (2018).
Van Woudenberg, N. et al. in Simulation, Image Processing, and Ultrasound Systems for Assisted Diagnosis and Navigation (eds Stoyanov, D. et al.) 74–81 (Springer, 2018).
Kusunose, K. et al. A deep learning approach for assessment of regional wall motion abnormality from echocardiographic images. JACC Cardiovasc. Imagin 13, 374–381 (2019).
Rajkomar, A. et al. Scalable and accurate deep learning with electronic health records. npj Digit. Med. 1, 18 (2018).
Kwon, J.-m, Kim, K.-H., Jeon, K.-H. & Park, J. Deep learning for predicting in-hospital mortality among heart disease patients based on echocardiography. Echocardiography 36, 213–218 (2019).
Avati, A. et al. Improving palliative care with deep learning. BMC Med. Inform. Decis. Mak. 18, 122 (2018).
Motwani, M. et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur. Heart J. 38, 500–507 (2016).
Hadamitzky, M. et al. Optimized prognostic score for coronary computed tomographic angiography: results from the confirm registry (coronary CT angiography evaluation for clinical outcomes: an international multicenter registry). J. Am. Coll. Cardiol. 62, 468–476 (2013).
Samad, M. D. et al. Predicting survival from large echocardiography and electronic health record datasets: optimization with machine learning. JACC Cardiovasc. Imaging 12, 681–689 (2018).
Jing, L. et al. A machine learning approach to management of heart failure populations. JACC Heart Fail. 8, 578–587 (2020).
Murillo, S. et al. Motion and deformation analysis of ultrasound videos with applications to classification of carotid artery plaques. In SPIE Medical Imaging (SPIE, 2012).
Cui, X. et al. Deformable regions of interest with multiple points for tissue tracking in echocardiography. Med. Image Anal. 35, 554–569 (2017).
Raghunath, S. et al. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat. Med. 26, 886–891 (2020).
Gahungu, N., Trueick, R., Bhat, S., Sengupta, P. P. & Dwivedi, G. Current challenges and recent updates in artificial intelligence and echocardiography. Curr. Cardiovasc. Imaging Rep. 13, 5 (2020).
Horgan, S. J. & Uretsky, S. in Essential Echocardiography: A Companion to Braunwald’s Heart Disease (eds Solomon, S. D. et al.) 460–473 (Elsevier, 2019).
Zhang, J. et al. Fully automated echocardiogram interpretation in clinical practice: feasibility and diagnostic accuracy. Circulation 138, 1623–1635 (2018).
Li, M. et al. Unified model for interpreting multi-view echocardiographic sequences without temporal information. Appl. Soft Comput. 88, 106049 (2020).
Ge, R. et al. K-net: Integrate left ventricle segmentation and direct quantification of paired echo sequence. IEEE Trans. Med. imaging 39, 1690–1702 (2019).
Ge, R. et al. Echoquan-net: direct quantification of echo sequence for left ventricle multidimensional indices via global-local learning, geometric adjustment and multi-target relation learning. In International Conference on Artificial Neural Networks (eds Tetko, I. et al.) 219–230 (Springer, 2019).
Jafari, M. H. et al. Automatic biplane left ventricular ejection fraction estimation with mobile point-of-care ultrasound using multi-task learning and adversarial training. Int. J. Comput. Assist. Radiol. Surg. 14, 1027–1037 (2019).
Ouyang, D. et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 580, 252–256 (2020).
Ghorbani, A. et al. Deep learning interpretation of echocardiograms. npj Digit. Med. 3, 10 (2020).
Behnami, D. et al. Automatic cine-based detection of patients at high risk of heart failure with reduced ejection fraction in echocardiograms. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. https://doi.org/10.1080/21681163.2019.1650398 (2019).
Yadlowsky, S. et al. Clinical implications of revised pooled cohort equations for estimating atherosclerotic cardiovascular disease risk. Ann. Intern. Med. 169, 20–29 (2018).
Levy, W. C. et al. The Seattle Heart Failure model. Circulation 113, 1424–1433 (2006).
McCarty, C. A. et al. The eMERGE network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics 4, 13 (2011).
Wehner, G. J. et al. Routinely reported ejection fraction and mortality in clinical practice: where does the nadir of risk lie? Eur. Heart J. 41, 1249–1257 (2020).
Liao, Z. et al. On modelling label uncertainty in deep neural networks: automatic estimation of intra-observer variability in 2D echocardiography quality assessment. IEEE Trans. Med. Imaging 39, 1868–1883 (2019).
Behnami, D. et al. Dual-view joint estimation of left ventricular ejection fraction with uncertainty modelling in echocardiograms. In International Conference on Medical Image Computing and Computer-Assisted Intervention (eds Shen, D. et al.) 696–704 (Springer, 2019).
Yancy, C. W. et al. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J. Am. Coll. Cardiol. 62, e147–e239 (2013).
Lund, L. H., Aaronson, K. D. & Mancini, D. M. Predicting survival in ambulatory patients with severe heart failure on beta-blocker therapy. Am. J. Cardiol. 92, 1350–1354 (2003).
Kavalieratos, D. et al. Palliative care in heart failure: rationale, evidence, and future priorities. J. Am. Coll. Cardiol. 70, 1919–1930 (2017).
Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 1, 206–215 (2019).
Arcadu, F. et al. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. npj Digit. Med. 2, 92 (2019).
Lee, H. et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat. Biomed. Eng. 3, 173 (2019).
Venugopalan, S. et al. Translating videos to natural language using deep recurrent neural networks. In Proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (Association for Computational Linguistics, 2015).
Madani, A., Arnaout, R., Mofrad, M. & Arnaout, R. Fast and accurate view classification of echocardiograms using deep learning. npj Digit. Med. 1, 6 (2018).
Tran, D., Bourdev, L., Fergus, R., Torresani, L. & Paluri, M. Learning spatiotemporal features with 3D convolutional networks. In Proc. IEEE International Conference on Computer Vision 4489–4497 (2015).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 2818–2826 (2016).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 4700–4708 (2017).
Prechelt, L. in Neural Networks: Tricks of the Trade (eds Montavon, G. et al.) 55–69 (Springer, 1998).
Buuren, S. & Groothuis-Oudshoorn, K. MICE: multivariate imputation by chained equations in R. J. Stat. Softw. 45, jss.v045.i03 (2011).
Chen, T. & Guestrin, C. Xgboost: a scalable tree boosting system. In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, 2016).
Williams, B. A. & Agarwal, S. Applying the Seattle Heart Failure model in the office setting in the era of electronic medical records. Circ. J. 82, 724–731 (2018).
Acknowledgements
This work was supported in part by funding from the Pennsylvania Dept of Health (SAP 4100070267 and 4100079720) and the Geisinger Health Plan and Clinic. The content of this article does not reflect the view of the funding sources.
Author information
Authors and Affiliations
Contributions
A.E.U.-C., C.M.H. and B.K.F. conceived the study and designed the experiments. A.E.U.-C. conducted all experiments. A.E.U.-C. and S.R. wrote the software for applying deep learning to echocardiography videos. A.E.U.-C., L.J., D.P.v., D.N.H., J.D.S. and J.B.L. assembled the input data. H.L.K., G.J.W., M.S.P. and A.A.P. gave advice on experiment design. L.J., C.D.N., C.M.H. and B.K.F. manually audited the data for the cardiologist survey. C.W.G., A.A., J.M.P. and B.J.C. completed the surveys and provided insights on interpretability experiments. A.E.U.-C., C.M.H., M.S.P. and B.K.F. wrote the manuscript. All authors critically revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
Geisinger receives funding from Tempus for ongoing development of predictive modelling technology and commercialization. Tempus and Geisinger have jointly applied for a patent related to this work. None of the authors has ownership interest in any of the intellectual property resulting from the partnership.
Additional information
Peer review information Nature Biomedical Engineering thanks Partho Sengupta, Purang Abolmaesumi and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, discussion, figures and tables.
Rights and permissions
About this article
Cite this article
Ulloa Cerna, A.E., Jing, L., Good, C.W. et al. Deep-learning-assisted analysis of echocardiographic videos improves predictions of all-cause mortality. Nat Biomed Eng 5, 546–554 (2021). https://doi.org/10.1038/s41551-020-00667-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-020-00667-9
This article is cited by
-
A comprehensive review on deep cardiovascular disease detection approaches: its datasets, image modalities and methods
Multimedia Tools and Applications (2024)
-
A deep learning-based electrocardiogram risk score for long term cardiovascular death and disease
npj Digital Medicine (2023)
-
Cartography of Genomic Interactions Enables Deep Analysis of Single-Cell Expression Data
Nature Communications (2023)
-
The Role of Artificial Intelligence in Echocardiography: A Clinical Update
Current Cardiology Reports (2023)
-
Generalizability and quality control of deep learning-based 2D echocardiography segmentation models in a large clinical dataset
The International Journal of Cardiovascular Imaging (2022)