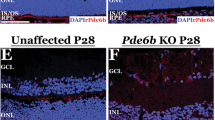
Abstract
Therapeutic genome editing requires effective and targeted delivery methods. The delivery of Cas9 mRNA using adeno-associated viruses has led to potent in vivo therapeutic efficacy, but can cause sustained Cas9 expression, anti-Cas9 immune responses and off-target edits. Lentiviral vectors have been engineered to deliver nucleases that are expressed transiently, but in vivo evidence of their biomedical efficacy is lacking. Here, we show that the lentiviral codelivery of Streptococcus pyogenes Cas9 mRNA and expression cassettes that encode a guide RNA that targets vascular endothelial growth factor A (Vegfa) is efficacious in a mouse model of wet age-related macular degeneration induced by Vegfa. A single subretinal injection of engineered lentiviruses knocked out 44% of Vegfa in retinal pigment epithelium and reduced the area of choroidal neovascularization by 63% without inducing off-target edits or anti-Cas9 immune responses. Engineered lentiviruses for the transient expression of nucleases may form the basis of new treatments for retinal neovascular diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Source data for the figures are available at Figshare (https://doi.org/10.6084/m9.figshare.12611819)76. The deep-sequencing and Nanopore DNA-sequencing data are available at the NCBI BioProject under the identifiers PRJNA642029, PRJNA593168 and PRJNA628164.
References
Dever, D. P. et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016).
Kim, K. et al. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res. 27, 419–426 (2017).
van Diemen, F. R. et al. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 12, e1005701 (2016).
Maeder, M. L. et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 25, 229–233 (2019).
Beyret, E. et al. Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson–Gilford progeria syndrome. Nat. Med. 25, 419–422 (2019).
Santiago-Fernandez, O. et al. Development of a CRISPR/Cas9-based therapy for Hutchinson–Gilford progeria syndrome. Nat. Med. 25, 423–426 (2019).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Li, A. et al. A self-deleting AAV-CRISPR system for in vivo genome editing. Mol. Ther. Methods Clin. Dev. 12, 111–122 (2019).
Chew, W. L. et al. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 13, 868–874 (2016).
Mingozzi, F. et al. Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci. Transl. Med. 5, 194ra192 (2013).
Nelson, C. E. et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 25, 427–432 (2019).
Wagner, D. L. et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 25, 242–248 (2019).
Wignakumar, T. & Fairchild, P. J. Evasion of pre-existing immunity to Cas9: a prerequisite for successful genome editing in vivo? Curr. Transplant. Rep. 6, 127–133 (2019).
Gao, X. et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217–221 (2018).
Montagna, C. et al. VSV-G-enveloped vesicles for traceless delivery of CRISPR-Cas9. Mol. Ther. Nucleic Acids 12, 453–462 (2018).
Lee, K. et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 1, 889–901 (2017).
Lee, B. et al. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2, 497–507 (2018).
Shahbazi, R. et al. Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nat. Mater. 18, 1124–1132 (2019).
Alkilany, A. M. & Murphy, C. J. Toxicity and cellular uptake of gold nanoparticles: what we have learned so far? J. Nanopart. Res. 12, 2313–2333 (2010).
Perry B Hackett, N. V. S. Gene therapy: delivering the second revolution in site-specific nucleases. eLife 3, e02904 (2014).
Cai, Y. & Mikkelsen, J. G. Lentiviral delivery of proteins for genome engineering. Curr. Gene Ther. 16, 194–206 (2016).
Naldini, L., Trono, D. & Verma, I. M. Lentiviral vectors, two decades later. Science 353, 1101–1102 (2016).
Cai, Y., Bak, R. O. & Mikkelsen, J. G. Targeted genome editing by lentiviral protein transduction of zinc-finger and TAL-effector nucleases. eLife 3, e01911 (2014).
Cai, Y. et al. DNA transposition by protein transduction of the PiggyBac transposase from lentiviral Gag precursors. Nucleic Acids Res. 42, e28 (2014).
Choi, J. G. et al. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 23, 627–633 (2016).
Lu, B. et al. Delivering SaCas9 mRNA by lentivirus-like bionanoparticles for transient expression and efficient genome editing. Nucleic Acids Res. 47, e44 (2019).
Lyu, P., Javidi-Parsijani, P., Atala, A. & Lu, B. Delivering Cas9/sgRNA ribonucleoprotein (RNP) by lentiviral capsid-based bionanoparticles for efficient ‘hit-and-run’ genome editing. Nucleic Acids Res. 47, e99 (2019).
Voelkel, C. et al. Protein transduction from retroviral Gag precursors. Proc. Natl Acad. Sci. USA 107, 7805–7810 (2010).
Prel, A. et al. Highly efficient in vitro and in vivo delivery of functional RNAs using new versatile MS2-chimeric retrovirus-like particles. Mol. Ther. Methods Clin. Dev. 2, 15039 (2015).
Mangeot, P. E. et al. Genome editing in primary cells and in vivo using viral-derived nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 10, 45 (2019).
Lim, L. S., Mitchell, P., Seddon, J. M., Holz, F. G. & Wong, T. Y. Age-related macular degeneration. Lancet 379, 1728–1738 (2012).
Andreoli, C. M. & Miller, J. W. Anti-vascular endothelial growth factor therapy for ocular neovascular disease. Curr. Opin. Ophthalmol. 18, 502–508 (2007).
Usui-Ouchi, A. & Friedlander, M. Anti-VEGF therapy: higher potency and long-lasting antagonism are not necessarily better. J. Clin. Invest. 129, 3032–3034 (2019).
Campochiaro, P. A. et al. Lentiviral vector gene transfer of endostatin/angiostatin for macular degeneration (GEM) study. Hum. Gene Ther. 28, 99–111 (2017).
Hughes, C. P. et al. AAV2/8 anti-angiogenic gene therapy using single-chain antibodies inhibits murine choroidal neovascularization. Mol. Ther. Methods Clin. Dev. 13, 86–98 (2019).
Murakami, Y. et al. Inhibition of choroidal neovascularization via brief subretinal exposure to a newly developed lentiviral vector pseudotyped with sendai viral envelope proteins. Hum. Gene Ther. 21, 199–209 (2009).
Ortinski, P. I., O’Donovan, B., Dong, X. & Kantor, B. Integrase-deficient lentiviral vector as an all-in-one platform for highly efficient CRISPR/Cas9-mediated gene editing. Mol. Ther. Methods Clin. Dev. 5, 153–164 (2017).
Sürün, D. et al. High efficiency gene correction in hematopoietic cells by donor-template-free CRISPR/Cas9 genome editing. Mol. Ther. Nucleic Acids 10, 1–8 (2018).
Holmgaard, A. et al. In vivo knockout of the Vegfa gene by lentiviral delivery of CRISPR/Cas9 in mouse retinal pigment epithelium cells. Mol. Ther. Nucleic Acids 9, 89–99 (2017).
Koo, T. et al. CRISPR-LbCpf1 prevents choroidal neovascularization in a mouse model of age-related macular degeneration. Nat. Commun. 9, 1855 (2018).
Saint-Geniez, M., Maldonado, A. E. & D’Amore, P. A. VEGF expression and receptor activation in the choroid during development and in the adult. Invest. Ophthalmol. Vis. Sci. 47, 3135–3142 (2006).
Kuzembayeva, M., Dilley, K., Sardo, L. & Hu, W. S. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455, 362–370 (2014).
Pickett, G. G. & Peabody, D. S. Encapsidation of heterologous RNAs by bacteriophage MS2 coat protein. Nucleic Acids Res. 21, 4621–4626 (1993).
Cai, Y. & Mikkelsen, J. G. Driving DNA transposition by lentiviral protein transduction. Mob. Genet. Elem. 4, e29591 (2014).
Skipper, K. A. et al. Time-restricted PiggyBac DNA transposition by transposase protein delivery using lentivirus-derived nanoparticles. Mol. Ther. Nucleic Acids 11, 253–262 (2018).
Ihry, R. J. et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Ellis, J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 16, 1241–1246 (2005).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Dang, Y. et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16, 280 (2015).
Wang, D. et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-Specific immune responses. Hum. Gene Ther. 26, 432–442 (2015).
Zeka, B. et al. Aquaporin 4-specific T cells and NMO-IgG cause primary retinal damage in experimental NMO/SD. Acta Neuropathol. Commun. 4, 82 (2016).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Gong, Y. et al. Optimization of an Image-guided laser-induced choroidal neovascularization model in mice. PLoS ONE 10, e0132643 (2015).
Staunstrup, N. H. et al. Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol. Ther. 17, 1205–1214 (2009).
Paludan, S. R., Reinert, L. S. & Hornung, V. DNA-stimulated cell death: implications for host defence, inflammatory diseases and cancer. Nat. Rev. Immunol. 19, 141–153 (2019).
Akcakaya, P. et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419 (2018).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR–Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Cheng, Y. & Tsai, S. Q. Illuminating the genome-wide activity of genome editors for safe and effective therapeutics. Genome Biol. 19, 226 (2018).
Stemmer, M., Thumberger, T., del Sol Keyer, M., Wittbrodt, J. & Mateo, J. L. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 10, e0124633 (2015).
Kurihara, T., Westenskow, P. D., Bravo, S., Aguilar, E. & Friedlander, M. Targeted deletion of Vegfa in adult mice induces vision loss. J. Clin. Invest. 122, 4213–4217 (2012).
Grishanin, R. et al. Preclinical evaluation of ADVM-022, a novel gene therapy approach to treating wet age-related macular degeneration. Mol. Ther. 27, 118–129 (2019).
Nishimura, T., Machida, S., Harada, T. & Kurosaka, D. Retinal ganglion cell function after repeated intravitreal injections of ranibizumab in patients with age-related macular degeneration. Clin. Ophthalmol. 6, 1073–1082 (2012).
Rakoczy, E. P. et al. Three-year follow-up of phase 1 and 2a rAAV.sFLT-1 subretinal gene therapy trials for exudative age-related macular degeneration. Am. J. Ophthalmol. 204, 113–123 (2019).
Rakoczy, E. P. et al. Gene therapy with recombinant adeno-associated vectors for neovascular age-related macular degeneration: 1 year follow-up of a phase 1 randomised clinical trial. Lancet 386, 2395–2403 (2015).
Campochiaro, P. A. Low risk to retina from sustained suppression of VEGF. J. Clin. Invest. 129, 3029–3031 (2019).
Richner, J. M. et al. Modified mRNA vaccines protect against Zika virus infection. Cell 168, 1114–1125 (2017).
Thuronyi, B. W. et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 37, 1070–1079 (2019).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846 (2018).
Park, J., Lim, K., Kim, J. S. & Bae, S. Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288 (2017).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Kluesner, M. G. et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J. 1, 239–250 (2018).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ling, S. et al. Dataset for Lentiviral delivery of co-packaged Cas9 mRNA and a Vegfa-targeting guide RNA prevents wet age-related macular degeneration in mice. Fighare https://doi.org/10.6084/m9.figshare.12611819 (2020).
Acknowledgements
We thank W. Yang at the Southern Medical University, China, for discussions and scientific input for the immunology part of the study. Y.C. is supported by the National Natural Science Foundation of China (no. 31971364), Pujiang Talent Project of Shanghai (no. 18PJ1404500), Shanghai Municipal Natural Science Foundation (no. 18ZR1419300), and startup funding from Shanghai Center for Systems Biomedicine, Shanghai Jiao Tong University (no. WF220441504).
Author information
Authors and Affiliations
Contributions
S.L. and Y.C. conceived the study and designed the experiments; S.L., S.Y., X.H., D.Y., Y.D., X.Q., D.W. and J.H. performed the experiments; all of the authors analysed the data; S.L. and Y.C. wrote the manuscript with help from all of the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures.
Rights and permissions
About this article
Cite this article
Ling, S., Yang, S., Hu, X. et al. Lentiviral delivery of co-packaged Cas9 mRNA and a Vegfa-targeting guide RNA prevents wet age-related macular degeneration in mice. Nat Biomed Eng 5, 144–156 (2021). https://doi.org/10.1038/s41551-020-00656-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-020-00656-y
This article is cited by
-
It takes two to tango with CRISPR: a history and overview of augmenting the technology for genetic engineering
Proceedings of the Indian National Science Academy (2024)
-
Recent advances in therapeutic CRISPR-Cas9 genome editing: mechanisms and applications
Molecular Biomedicine (2023)
-
The application and progression of CRISPR/Cas9 technology in ophthalmological diseases
Eye (2023)
-
CRISPR/Cas9 mediated specific ablation of vegfa in retinal pigment epithelium efficiently regresses choroidal neovascularization
Scientific Reports (2023)
-
Advances in CRISPR therapeutics
Nature Reviews Nephrology (2023)