Abstract
Engineered human mini-brains, made possible by knowledge from the convergence of precision microengineering and cell biology, permit systematic studies of complex neurological processes and of pathogenesis beyond what can be done with animal models. By culturing human brain cells with physiological microenvironmental cues, human mini-brain models reconstitute the arrangement of structural tissues and some of the complex biological functions of the human brain. In this Review, we highlight the most significant developments that have led to microphysiological human mini-brain models. We introduce the history of mini-brain development, review methods for creating mini-brain models in static conditions, and discuss relevant state-of-the-art dynamic cell-culture systems. We also review human mini-brain models that reconstruct aspects of major neurological disorders under static or dynamic conditions. Engineered human mini-brains will contribute to advancing the study of the physiology and aetiology of neurological disorders, and to the development of personalized medicines for them.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

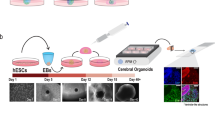
Image courtesy of You Jung Kang, University of North Carolina at Charlotte



Similar content being viewed by others
References
Wyss-Coray, T. Ageing, neurodegeneration and brain rejuvenation. Nature 539, 180–186 (2016).
Abbott, N. J., Patabendige, A. A. K., Dolman, D. E. M., Yusof, S. R. & Begley, D. J. Structure and function of the blood–brain barrier. Neurobiol. Dis. 37, 13–25 (2010).
Zlokovic, B. V. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat. Rev. Neurosci. 12, 723–738 (2011).
Sturchler-Pierrat, C. et al. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc. Natl Acad. Sci. USA 94, 13287–13292 (1997).
Greek, R. & Menache, A. Systematic reviews of animal models: methodology versus epistemology. Int. J. Med. Sci. 10, 206–221 (2013).
Ferdowsian, H. R. & Beck, N. Ethical and scientific considerations regarding animal testing and research. PLoS ONE 6, e24059 (2011).
Harrison, R. G. The outgrowth of the nerve fiber as a mode of protoplasmic movement. J. Exp. Zool. 9, 787–846 (1910).
Han, D. W. et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10, 465–472 (2012).
Vierbuchen, T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010).
Wang, P. et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism 8, 11 (2017).
Hardy, J. & Allsop, D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 12, 383–388 (1991).
Maries, E., Dass, B., Collier, T. J., Kordower, J. H. & Steece-Collier, K. The role of α-synuclein in Parkinson’s disease: insights from animal models. Nat. Rev. Neurosci. 4, 727–738 (2003).
Visanji, N. P., Brooks, P. L., Hazrati, L. N. & Lang, A. E. The prion hypothesis in Parkinson’s disease: Braak to the future. Acta Neuropathol. Commun. 1, 2 (2013).
Brundin, P. & Melki, R. Prying into the prion hypothesis for Parkinson’s disease. J. Neurosci. 37, 9808–9818 (2017).
Booth, R. & Kim, H. Characterization of a microfluidic in vitro model of the blood–brain barrier (μBBB). Lab Chip 12, 1784–1792 (2012).
Cucullo, L. et al. Immortalized human brain endothelial cells and flow-based vascular modeling: a marriage of convenience for rational neurovascular studies. J. Cereb. Blood Flow Metab. 28, 312–328 (2008).
Booth, R., Noh, S. & Kim, H. A multiple-channel, multiple-assay platform for characterization of full-range shear stress effects on vascular endothelial cells. Lab Chip 14, 1880–1890 (2014).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Zhang, S., Wernig, M., Duncan, I. D. & Thomson, J. A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nature 19, 1129–1133 (2001).
Campenot, R. B. Local control of neurite development by nerve growth factor. Proc. Natl Acad. Sci. USA 74, 4516–4519 (1977).
Rubin, L. L. et al. A cell culture model of the blood–brain barrier. J. Cell Biol. 115, 1725–1735 (1991).
Patabendige, A., Skinner, R. A., Morgan, L. & Abbott, N. J. A detailed method for preparation of a functional and flexible blood–brain barrier model using porcine brain endothelial cells. Brain Res. 1521, 16–30 (2013).
Patabendige, A., Skinner, R. A. & Abbott, N. J. Establishment of a simplified in vitro porcine blood–brain barrier model with high transendothelial electrical resistance. Brain Res. 1521, 1–15 (2013).
Hatherell, K., Couraud, P. O., Romero, I. A., Weksler, B. & Pilkington, G. J. Development of a three-dimensional, all-human in vitro model of the blood–brain barrier using mono-, co-, and tri-cultivation Transwell models. J. Neurosci. Methods 199, 223–229 (2011).
Freese, C. et al. A novel blood–brain barrier co-culture system for drug targeting of Alzheimer’s disease: establishment by using acitretin as a model drug. PLoS ONE 9, e91003 (2014).
Appelt-Menzel, A. et al. Establishment of a human blood–brain-barrier co-culture model mimicking the neurovascular unit using induced pluri- and multipotent stem cells. Stem Cell Rep. 8, 894–906 (2017).
Al-Shehri, A. et al. Permeability of PEGylated immunoarsonoliposomes through in vitro blood brain barrier-medulloblastoma co-culture models for brain tumor therapy. Pharm. Res. 32, 1072–1083 (2015).
Biernacki, K., Prat, A., Blain, M. & Antel, J. P. Regulation of Th1 and Th2 lymphocyte migration by human adult brain endothelial cells. J. Neuropathol. Exp. Neurol. 60, 1127–1136 (2001).
Thomsen, L. B., Burkhart, A. & Moos, T. A triple culture model of the blood–brain barrier using porcine brain endothelial cells, astrocytes and pericytes. PLoS ONE 10, e0134765 (2015).
Johnsen, K. B. et al. Targeting transferrin receptors at the blood–brain barrier improves the uptake of immunoliposomes and subsequent cargo transport into the brain parenchyma. Sci. Rep. 7, 10396 (2017).
Ruoslahti, E. Brain extracellular matrix. Glycobiology 6, 489–492 (1996).
Dana, H. et al. Hybrid multiphoton volumetric functional imaging of large-scale bioengineered neuronal networks. Nat. Commun. 5, 3997 (2014).
Thomson, D. Controlled growth en masse (somatic growth) of embryonic chick tissue in vitro. Proc. R. Soc. Med. 7, 71–75 (1914).
Aimetti, A. A., Machen, A. J. & Anseth, K. S. Poly(ethylene glycol) hydrogels formed by thiol-ene photopolymerization for enzyme-responsive protein delivery. Biomaterials 30, 6048–6054 (2009).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Reynolds, B. A., Tetzlaff, W. & Weiss, S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J. Neurosci. 12, 4565–4574 (1992).
Eiraku, M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532 (2008).
Boutin, M. E. et al. A three-dimensional neural spheroid model for capillary-like network formation. J. Neurosci. Methods 299, 55–63 (2016).
Dingle, Y.-T. L. et al. Three-dimensional neural spheroid culture: an in vitro model for cortical studies. Tissue Eng. Part C Methods 21, 1274–1283 (2015).
Jo, J. et al. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 19, 248–257 (2016).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA 112, 15672–15677 (2015).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Velasco, S. et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019).
Lancaster, M. A. et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 35, 659–666 (2017).
Li, Y. et al. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 20, 385–396 (2017).
Ogawa, J., Pao, G. M., Shokhirev, M. N. & Verma, I. M. Glioblastoma model using human cerebral organoids. Cell Rep. 23, 1220–1229 (2018).
Abbott, N. J. Blood–brain barrier structure and function and the challenges for CNS drug delivery. J. Inherit. Metab. Dis. 36, 437–449 (2013).
Cui, H., Nowicki, M., Fisher, J. P. & Zhang, L. G. 3D Bioprinting for organ regeneration. Adv. Healthc. Mater. 6, 1601118 (2017).
Murphy, S. V. & Atala, A. 3D bioprinting of tissues and organs. Nat. Biotechnol. 32, 773–785 (2014).
Ravnic, D. J. et al. Transplantation of bioprinted tissues and organs technical and clinical challenges and future perspectives. Ann. Surg. 266, 48–58 (2017).
Li, J., Chen, M., Fan, X. & Zhou, H. Recent advances in bioprinting techniques: approaches, applications and future prospects. J. Transl. Med. 14, 271 (2016).
Sanjana, N. E. & Fuller, S. B. A fast flexible ink-jet printing method for patterning dissociated neurons in culture. J. Neurosci. Methods 136, 151–163 (2004).
Xu, T. et al. Viability and electrophysiology of neural cell structures generated by the inkjet printing method. Biomaterials 27, 3580–3588 (2006).
Lee, Y. B. et al. Bio-printing of collagen and VEGF-releasing fibrin gel scaffolds for neural stem cell culture. Exp. Neurol. 223, 645–652 (2010).
Lee, W. et al. Three-dimensional bioprinting of rat embryonic neural cells. NeuroReport 20, 798–803 (2009).
Patz, T. M. et al. Three-dimensional direct writing of B35 neuronal cells. J. Biomed. Mater. Res. Part B Appl. Biomater. 78, 124–130 (2006).
Lozano, R. et al. 3D printing of layered brain-like structures using peptide modified gellan gum substrates. Biomaterials 67, 264–273 (2015).
Hsieh, F. Y., Lin, H. H. & Hsu, S. 3D bioprinting of neural stem cell-laden thermoresponsive biodegradable polyurethane hydrogel and potential in central nervous system repair. Biomaterials 71, 48–57 (2015).
Curley, J. L. et al. Isolated node engineering of neuronal systems using laser direct write. Biofabrication 8, 015013 (2016).
Heinrich, M. A. et al. 3D-bioprinted mini-brain: a glioblastoma model to study cellular interactions and therapeutics. Adv. Mater. 31, e1806590 (2019).
Gu, Q. et al. Functional 3D neural mini-tissues from printed gel-based bioink and human neural stem cells. Adv. Healthc. Mater. 5, 1429–1438 (2016).
Abbott, N. J., Rönnbäck, L. & Hansson, E. Astrocyte–endothelial interactions at the blood–brain barrier. Nat. Rev. Neurosci. 7, 41–53 (2006).
Alrifaiy, A., Lindahl, O. A. & Ramser, K. Polymer-based microfluidic devices for pharmacy, biology and tissue engineering. Polymers (Basel) 4, 1349–1398 (2012).
Bhatia, S. N. & Ingber, D. E. Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772 (2014).
Duffy, D. C., McDonald, J. C., Schueller, O. J. A. & Whitesides, G. M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Toepke, M. W. & Beebe, D. J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 6, 1484–1486 (2006).
Chumbimuni-Torres, K. Y. et al. Adsorption of proteins to thin-films of PDMS and its effect on the adhesion of human endothelial cells. RSC Adv. 1, 706–714 (2011).
Haring, A. P., Sontheimer, H. & Johnson, B. N. Microphysiological human brain and neural systems-on-a-chip: potential alternatives to small animal models and emerging platforms for drug discovery and personalized medicine. Stem Cell Rev. Rep. 13, 381–406 (2017).
Taylor, A. M. et al. Microfluidic multicompartment device for neuroscience research. Langmuir 19, 1551–1556 (2003).
Taylor, A. M. et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods 2, 599–605 (2005).
Yang, I. H., Siddique, R., Hosmane, S., Thakor, N. & Höke, A. Compartmentalized microfluidic culture platform to study mechanism of paclitaxel-induced axonal degeneration. Exp. Neurol. 218, 124–128 (2009).
Samson, A. J., Robertson, G., Zagnoni, M. & Connolly, C. N. Neuronal networks provide rapid neuroprotection against spreading toxicity. Sci. Rep. 6, 33746 (2016).
Taylor, A. M., Dieterich, D. C., Ito, H. T., Kim, S. A. & Schuman, E. M. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron 66, 57–68 (2010).
Hengst, U., Deglincerti, A., Kim, H. J., Jeon, N. L. & Jaffrey, S. R. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat. Cell Biol. 11, 1024–1030 (2009).
Rivieccio, M. A. et al. HDAC6 is a target for protection and regeneration following injury in the nervous system. Proc. Natl Acad. Sci. USA 106, 19599–19604 (2009).
Kilic, O. et al. Brain-on-a-chip model enables analysis of human neuronal differentiation and chemotaxis. Lab Chip 16, 4152–4162 (2016).
Wang, J. D., Khafagy, E. S., Khanafer, K., Takayama, S. & Elsayed, M. E. H. Organization of endothelial cells, pericytes, and astrocytes into a 3D microfluidic in vitro model of the blood–brain barrier. Mol. Pharm. 13, 895–906 (2016).
Achyuta, A. K. H. et al. A modular approach to create a neurovascular unit-on-a-chip. Lab Chip 13, 542–553 (2013).
Griep, L. M. et al. BBB ON CHIP: microfluidic platform to mechanically and biochemically modulate blood–brain barrier function. Biomed. Microdevices 15, 145–150 (2013).
Cho, H. et al. Three-dimensional blood–brain barrier model for in vitro studies of neurovascular pathology. Sci. Rep. 5, 15222 (2015).
Xu, H. et al. A dynamic in vivo-like organotypic blood–brain barrier model to probe metastatic brain tumors. Sci. Rep. 6, 36670 (2016).
Adriani, G., Ma, D., Pavesi, A. & Kamm, R. D. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood–brain barrier. Lab Chip 12, 169–182 (2017).
Bang, S. et al. A low permeability microfluidic blood–brain barrier platform with direct contact between perfusable vascular network and astrocytes. Sci. Rep. 7, 8083 (2017).
Brown, J. A. et al. Recreating blood–brain barrier physiology and structure on chip: a novel neurovascular microfluidic bioreactor. Biomicrofluidics 9, 054124 (2015).
Herland, A. et al. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood–brain barrier on a chip. PLoS ONE 11, e0150360 (2016).
Park, T. E. et al. Hypoxia-enhanced Blood–Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 10, 2621 (2019).
Walter, F. R. et al. A versatile lab-on-a-chip tool for modeling biological barriers. Sens. Actuators B Chem. 222, 1209–1219 (2016).
Wang, Y. I., Abaci, H. E. & Shuler, M. L. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol. Bioeng. 114, 184–194 (2017).
Seidel, D., Jahnke, H.-G., Englich, B., Girard, M. & Robitzki, A. A. In vitro field potential monitoring on a multi-microelectrode array for the electrophysiological long-term screening of neural stem cell maturation. Analyst 142, 1929–1937 (2017).
Hofmann, F. & Bading, H. Long term recordings with microelectrode arrays: studies of transcription-dependent neuronal plasticity and axonal regeneration. J. Physiol. Paris 99, 125–132 (2006).
Fan, Y., Nguyen, D. T., Akay, Y., Xu, F. & Akay, M. Engineering a brain cancer chip for high-throughput drug screening. Sci. Rep. 6, 25062 (2016).
Campisi, M. et al. 3D self-organized microvascular model of the human blood–brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials 180, 117–129 (2018).
Park, J. et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 21, 941–951 (2018).
Osaki, T., Sivathanu, V. & Kamm, R. D. Engineered 3D vascular and neuronal networks in a microfluidic platform. Sci. Rep. 8, 5168 (2018).
Lee, S. R. et al. Modeling neural circuit, blood–brain barrier, and myelination on a microfluidic 96 well plate. Biofabrication 11, 035013 (2019).
Moreno, E. L. et al. Differentiation of neuroepithelial stem cells into functional dopaminergic neurons in 3D microfluidic cell culture. Lab Chip 15, 2419–2428 (2015).
Wevers, N. R. et al. High-throughput compound evaluation on 3D networks of neurons and glia in a microfluidic platform. Sci. Rep. 6, 38856 (2016).
Shi, P. et al. Synapse microarray identification of small molecules that enhance synaptogenesis. Nat. Commun. 2, 510 (2011).
Lee, J. N., Park, C. & Whitesides, G. M. Solvent compatibility of poly(dimethylsiloxane)-based microfluidic devices. Anal. Chem. 75, 6544–6554 (2003).
Riaz, A. et al. Reactive deposition of nano-films in deep polymeric microcavities. Lab Chip 12, 4877 (2012).
Abbott, N. J. Evidence for bulk flow of brain interstitial fluid: significance for physiology and pathology. Neurochem. Int. 45, 545–552 (2004).
Park, J. et al. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of Alzheimer’s disease. Lab Chip 15, 141–150 (2014).
Choi, Y. J. et al. Neurotoxic amyloid beta oligomeric assemblies recreated in microfluidic platform with interstitial level of slow flow. Sci. Rep. 3, 1921 (2013).
Wong, A. D. et al. The blood–brain barrier: an engineering perspective. Front. Neuroeng. 6, 7 (2013).
Stanness, K. A. et al. Morphological and functional characterization of an in vitro blood–brain barrier model. Brain Res. 771, 329–342 (1997).
Stanness, K. A., Guatteo, E. & Janigro, D. A dynamic model of the blood–brain barrier in vitro. Neurotoxicology 17, 481–496 (1996).
Sei, Y., Justus, K., LeDuc, P. & Kim, Y. Engineering living systems on chips: from cells to human on chips. Microfluid. Nanofluid. 16, 907–920 (2014).
Vatine, G. D. et al. Human iPSC-derived blood–brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 24, 995–1005 (2019).
Wang, Y., Wang, L., Guo, Y., Zhu, Y. & Qin, J. Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. RSC Adv. 8, 1677–1685 (2018).
Cakir, B. et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 16, 1169–1175 (2019).
Berdichevsky, Y., Staley, K. J. & Yarmush, M. L. Building and manipulating neural pathways with microfluidics. Lab Chip 10, 999–1004 (2010).
Scott, A. et al. A microfluidic microelectrode array for simultaneous electrophysiology, chemical stimulation, and imaging of brain slices. Lab Chip 13, 527–535 (2013).
Hardy, J. & Selkoe, D. J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002).
Choi, S. H. et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 515, 274–278 (2014).
Lee, H.-K. et al. Three dimensional human neuro-spheroid model of Alzheimer’s disease based on differentiated induced pluripotent stem cells. PLoS ONE 11, e0163072 (2016).
Raja, W. K. et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE 11, e0161969 (2016).
Winklhofer, K. F. & Haass, C. Mitochondrial dysfunction in Parkinson’s disease. Biochim. Biophys. 1802, 29–44 (2010).
Song, H. L. et al. β-Amyloid is transmitted via neuronal connections along axonal membranes. Ann. Neurol. 75, 88–97 (2014).
Deleglise, B. et al. β-Amyloid induces a dying-back process and remote trans-synaptic alterations in a microfluidic-based reconstructed neuronal network. Acta Neuropathol. Commun. 2, 145 (2014).
Poon, W. W. et al. β-Amyloid impairs axonal BDNF retrograde trafficking. Neurobiol. Aging 32, 821–833 (2011).
Cho, H. et al. Microfluidic chemotaxis platform for differentiating the roles of soluble and bound amyloid-β on microglial accumulation. Sci. Rep. 3, 1823 (2013).
Dujardin, S. et al. Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: relevance to sporadic tauopathies. Acta Neuropathol. Commun. 2, 14 (2014).
Calafate, S. et al. Synaptic contacts enhance cell-to-cell Tau pathology propagation. Cell Rep. 11, 1176–1183 (2015).
Takeda, S. et al. Neuronal uptake and propagation of a rare phosphorylated high-molecular-weight tau derived from Alzheimer’s disease brain. Nat. Commun. 6, 8490 (2015).
Wang, Y. et al. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener. 12, 5 (2017).
Iliff, J. J. et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 4, 147ra111 (2012).
Zhang, W. et al. Neuromelanin activates microglia and induces degeneration of dopaminergic neurons: implications for progression of Parkinson’s disease. Neurotox. Res. 19, 63–72 (2011).
Zecca, L., Zucca, F. A., Wilms, H. & Sulzer, D. Neuromelanin of the substantia nigra: a neuronal black hole with protective and toxic characteristics. Trends Neurosci. 26, 578–580 (2003).
Lee, H. J. et al. Dopamine promotes formation and secretion of non-fibrillar alpha-synuclein oligomers. Exp. Mol. Med. 43, 216–222 (2011).
Volpicelli-Daley, L. A. et al. Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72, 57–71 (2011).
Hu, Q. & Wang, G. Mitochondrial dysfunction in Parkinson’s disease. Transl. Neurodegener. 5, 14 (2016).
Lu, X., Kim-Han, J. S., Malley, K. L. O. & Sakiyama-Elbert, S. E. A microdevice platform for visualizing mitochondrial transport in aligned dopaminergic axons. J. Neurosci. Methods 209, 35–39 (2012).
Huse, J. T. & Holland, E. C. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat. Rev. Cancer 10, 319–331 (2010).
Chonan, Y., Taki, S., Sampetrean, O., Saya, H. & Sudo, R. Endothelium-induced three-dimensional invasion of heterogeneous glioma initiating cells in a microfluidic coculture platform. Integr. Biol. 9, 762–773 (2017).
Beauchesne, P. Extra-neural metastases of malignant gliomas: myth or reality? Cancers (Basel) 3, 461–477 (2011).
Inglese, M. et al. Diffuse axonal injury in mild traumatic brain injury: a diffusion tensor imaging study. J. Neurosurg. 103, 298–303 (2005).
Zetterberg, H., Smith, D. H. & Blennow, K. Biomarkers of mild traumatic brain injury in cerebrospinal fluid and blood. Nat. Rev. Neurol. 9, 201–210 (2013).
Sweeney, M. D., Sagare, A. P. & Zlokovic, B. V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133–150 (2018).
Johnson, V. E. et al. Mechanical disruption of the blood–brain barrier following experimental concussion. Acta Neuropathol. 135, 711–726 (2018).
Bar-Kochba, E., Scimone, M. T., Estrada, J. B. & Franck, C. Strain and rate-dependent neuronal injury in a 3D in vitro compression model of traumatic brain injury. Sci. Rep. 6, 30550 (2016).
Tang-Schomer, M. D. et al. Bioengineered functional brain-like cortical tissue. Proc. Natl Acad. Sci. USA 111, 13811–13816 (2014).
Kanemaru, K. et al. Calcium-dependent N-cadherin up-regulation mediates reactive astrogliosis and neuroprotection after brain injury. Proc. Natl Acad. Sci. USA 110, 11612–11617 (2013).
Madathil, S. K. et al. Astrocyte-specific overexpression of insulin-like growth factor-1 protects hippocampal neurons and reduces behavioral deficits following traumatic brain injury in mice. PLoS ONE 8, e67204 (2013).
Moon, L. D. F. & Fawcett, J. W. Reduction in CNS scar formation without concomitant increase in axon regeneration following treatment of adult rat brain with a combination of antibodies to TGFβ1 and β2. Eur. J. Neurosci. 14, 1667–1677 (2001).
Roth, T. L. et al. Transcranial amelioration of inflammation and cell death after brain injury. Nature 505, 223–228 (2014).
Schachtrup, C. et al. Fibrinogen triggers astrocyte scar formation by promoting the availability of active TGF-β after vascular damage. J. Neurosci. 30, 5843–5854 (2010).
Gorina, R., Font-Nieves, M., Márquez-Kisinousky, L., Santalucia, T. & Planas, A. M. Astrocyte TLR4 activation induces a proinflammatory environment through the interplay between MyD88-dependent NFκB signaling, MAPK, and Jak1/Stat1 pathways. Glia 59, 242–255 (2011).
Pedrazzi, M. et al. Selective proinflammatory activation of astrocytes by high mobility group Box 1 protein signaling. J. Immunol. 179, 8525–8532 (2007).
Ponath, G. et al. Autocrine S100B effects on astrocytes are mediated via RAGE. J. Neuroimmunol. 184, 214–222 (2007).
Maneshi, M. M., Sachs, F. & Hua, S. Z. A threshold shear force for calcium influx in an astrocyte model of traumatic brain injury. J. Neurotrauma 32, 1020–1029 (2015).
Mansour, A. A. et al. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432–441 (2018).
Ehsan, S. M., Welch-Reardon, K. M., Waterman, M. L., Hughes, C. C. W. & George, S. C. A three-dimensional in vitro model of tumor cell intravasation. Integr. Biol. 6, 603–610 (2014).
Wimmer, R. A. et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019).
Kim, S. et al. Transneuronal propagation of pathologic α-synuclein from the gut to the brain models Parkinson’s disease. Neuron 103, 627–641 (2019).
Mass, E. et al. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature 549, 389–393 (2017).
Zhang, W. et al. Aggregated α-synuclein activates microglia: a process leading to disease progression in Parkinson’s disease. FASEB J. 19, 533–542 (2005).
Tang, T.-S. et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc. Natl Acad. Sci. USA 102, 2602–2607 (2005).
Presgraves, S. P., Ahmed, T., Borwege, S. & Joyce, J. N. Terminally differentiated SH-SY5Y cells provide a model system for studying neuroprotective effects of dopamine agonists. Neurotox. Res. 5, 579–598 (2004).
Domert, J. et al. Neurobiology of disease spreading of amyloid-β peptides via neuritic cell-to-cell transfer is dependent on insufficient cellular clearance. Neurobiol. Dis. 65, 82–92 (2014).
An, M. C. et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell 11, 253–263 (2012).
Dai, X., Ma, C., Lan, Q. & Xu, T. 3D bioprinted glioma stem cells for brain tumor model and applications of drug susceptibility. Biofabrication 8, 045005 (2016).
Dai, X. et al. Coaxial 3D bioprinting of self-assembled multicellular heterogeneous tumor fibers. Sci. Rep. 7, 1457 (2017).
Terrell-Hall, T. B., Ammer, A. G., Griffith, J. I. G. & Lockman, P. R. Permeability across a novel microfluidic blood–tumor barrier model. Fluids Barriers CNS 14, 3 (2017).
Prabhakarpandian, B. et al. SyM-BBB: a microfluidic blood brain barrier model. Lab Chip 13, 1093–1101 (2013).
Maoz, B. M. et al. Organs-on-chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 17, 2294–2302 (2017).
Osaki, T., Uzel, S. G. M. & Kamm, R. D. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 4, eaat5847 (2018).
Shao, J. et al. Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress. Lab Chip 9, 3118–3125 (2009).
Yap, Y. C. et al. Mild and repetitive very mild axonal stretch injury triggers cystoskeletal mislocalization and growth cone collapse. PLoS ONE 12, e0176997 (2017).
Hosmane, S. et al. Toll/interleukin-1 receptor domain-containing adapter inducing interferon-β mediates microglial phagocytosis of degenerating axons. J. Neurosci. 32, 7745–7757 (2012).
Tang-Schomer, M. D., Davies, P., Graziano, D., Thurber, A. E. & Kaplan, D. L. Neural circuits with long-distance axon tracts for determining functional connectivity. J. Neurosci. Methods 222, 82–90 (2014).
Seidi, A. et al. A microfluidic-based neurotoxin concentration gradient for the generation of an in vitro model of Parkinson’s disease. Biomicrofluidics 5, 22214 (2011).
Dollé, J.-P., Morrison, B. III, Schloss, R. S. & Yarmush, M. L. An organotypic uniaxial strain model using microfluidics. Lab Chip 13, 432–442 (2013).
Tang, Y. T., Kim, J., Lopez-Valdes, H. E., Brennan, K. C. & Ju, Y. S. Microfluidic chamber with active suction ports for localized chemical stimulation of brain slices. Lab Chip 11, 2247–2254 (2011).
Karzbrun, E., Kshirsagar, A., Cohen, S. R., Hanna, J. H. & Reiner, O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 14, 515–522 (2018).
Sei, Y. J., Ahn, S. I., Virtue, T., Kim, T. & Kim, Y. Detection of frequency-dependent endothelial response to oscillatory shear stress using a microfluidic transcellular monitor. Sci. Rep. 7, 10019 (2017).
Iadecola, C. & Davisson, R. L. Hypertension and cerebrovascular dysfunction. Cell Metab. 7, 476–484 (2008).
Lee, R. T. & Kamm, R. D. Vascular mechanics for the cardiologist. J. Am. Coll. Cardiol. 23, 1289–1295 (1994).
Zhao, Z., Nelson, A. R., Betsholtz, C. & Zlokovic, B. V. Establishment and dysfunction of the blood–brain barrier. Cell 163, 1064–1078 (2015).
Cunningham, C., Dunne, A. & Lopez-Rodriguez, A. B. Astrocytes: heterogeneous and dynamic phenotypes in neurodegeneration and innate immunity. Neuroscientist 25, 455–474 (2018).
Hernandez-Ontiveros, D. G. et al. Microglia activation as a biomarker for traumatic brain injury. Front. Neurol. 4, 30 (2013).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Napoli, I. & Neumann, H. Microglial clearance function in health and disease. Neuroscience 158, 1030–1038 (2009).
Acknowledgements
The authors thank W.-B. Tay (Ministry of Education, Singapore) and S. Neo (National University of Singapore) for proofreading the manuscript. This work was supported by the National Research Foundation (nos. NRF-2020R1A2C2010285 and NRF-2018M3C7A1056896 to H.C.)
Author information
Authors and Affiliations
Contributions
L.P.L. and H.C. shaped ideas and provided guidance. H.Y.T. and H.C. researched data for the article. H.Y.T. prepared the figures and wrote the manuscript with assistance from H.C. and L.P.L. All authors contributed to discussion of the contents, and reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tan, HY., Cho, H. & Lee, L.P. Human mini-brain models. Nat Biomed Eng 5, 11–25 (2021). https://doi.org/10.1038/s41551-020-00643-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-020-00643-3
This article is cited by
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
Environmental aluminum oxide inducing neurodegeneration in human neurovascular unit with immunity
Scientific Reports (2024)
-
Three-dimensional human neural culture on a chip recapitulating neuroinflammation and neurodegeneration
Nature Protocols (2023)
-
Functional bioengineered models of the central nervous system
Nature Reviews Bioengineering (2023)
-
Phenotypic, metabolic, and biogenesis properties of human stem cell-derived cerebellar spheroids
Scientific Reports (2022)