Abstract
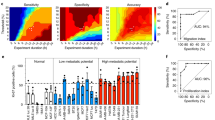
Clinical scores, molecular markers and cellular phenotypes have been used to predict the clinical outcomes of patients with glioblastoma. However, their clinical use has been hampered by confounders such as patient co-morbidities, by the tumoral heterogeneity of molecular and cellular markers, and by the complexity and cost of high-throughput single-cell analysis. Here, we show that a microfluidic assay for the quantification of cell migration and proliferation can categorize patients with glioblastoma according to progression-free survival. We quantified with a composite score the ability of primary glioblastoma cells to proliferate (via the protein biomarker Ki-67) and to squeeze through microfluidic channels, mimicking aspects of the tight perivascular conduits and white-matter tracts in brain parenchyma. The assay retrospectively categorized 28 patients according to progression-free survival (short-term or long-term) with an accuracy of 86%, predicted time to recurrence and correctly categorized five additional patients on the basis of survival prospectively. RNA sequencing of the highly motile cells revealed differentially expressed genes that correlated with poor prognosis. Our findings suggest that cell-migration and proliferation levels can predict patient-specific clinical outcomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, but they are available for research purposes from the corresponding authors on reasonable request. RNA-seq data are available at the National Center for Biotechnology Information Gene Expression Omnibus under accession no. GSE144610.
References
Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 20 (Suppl. 4), iv1–iv86 (2018).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Chaichana, K. L. et al. Establishing percent resection and residual volume thresholds affecting survival and recurrence for patients with newly diagnosed intracranial glioblastoma. Neuro Oncol. 16, 113–122 (2014).
Shah, S. R. et al. YAP controls cell migration and invasion through a Rho-GTPase switch. Preprint at https://doi.org/10.1101/602052 (2019).
Chaichana, K. L. et al. Multiple resections for patients with glioblastoma: prolonging survival. J. Neurosurg. 118, 812–820 (2013).
Chaichana, K., Parker, S., Olivi, A. & Quinones-Hinojosa, A. A proposed classification system that projects outcomes based on preoperative variables for adult patients with glioblastoma multiforme. J. Neurosurg. 112, 997–1004 (2010).
Wei, S. et al. Heterozygous IDH1R132H/WT created by ‘single base editing’ inhibits human astroglial cell growth by downregulating YAP. Oncogene 37, 5160–5174 (2018).
Grossman, R. et al. MGMT inactivation and clinical response in newly diagnosed GBM patients treated with Gliadel. J. Clin. Neurosci. 22, 1938–1942 (2015).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Noushmehr, H. et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 (2010).
Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006).
Verhaak, R. G. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell 17, 98–110 (2010).
Colman, H. & Aldape, K. Molecular predictors in glioblastoma: toward personalized therapy. Arch. Neurol. 65, 877–883 (2008).
Jaeckle, K. A. et al. Correlation of tumor O6 methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: a Southwest Oncology Group study. J. Clin. Oncol. 16, 3310–3315 (1998).
Hegi, M. E. et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 10, 1871–1874 (2004).
Hegi, M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 (2005).
Shah, S. R., Quinones-Hinojosa, A. & Xia, S. Advances in brain cancer: creating monoallelic single point mutation in IDH1 by single base editing. J. Oncol. Res. Ther. 5, 166 (2018).
Preusser, M. et al. Anti-O6-methylguanine-methyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 18, 520–532 (2008).
Grasbon-Frodl, E. M. et al. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J. Cancer 121, 2458–2464 (2007).
Hartmann, C. et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 118, 469–474 (2009).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009).
Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 372, 2481–2498 (2015).
Eckel-Passow, J. E. et al. Glioma groups based on 1p/19q, IDH and TERT promoter mutations in tumors. N. Engl. J. Med. 372, 2499–2508 (2015).
Joo, K. M. et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 3, 260–273 (2013).
Specht, H. & Slavov, N. Transformative opportunities for single-cell proteomics. J. Proteome Res. 17, 2565–2571 (2018).
Wills, Q. F. & Mead, A. J. Application of single-cell genomics in cancer: promise and challenges. Hum. Mol. Genet. 24, R74–R84 (2015).
Chandler, Y. et al. Cost effectiveness of gene expression profile testing in community practice. J. Clin. Oncol. 36, 554–562 (2018).
Lippman, M. & Osborne, C. K. Circulating tumor DNA—ready for prime time? N. Engl. J. Med. 368, 1249–1250 (2013).
Smith, C. L. et al. Pre-exposure of human adipose mesenchymal stem cells to soluble factors enhances their homing to brain cancer. Stem Cells Transl. Med. 4, 239–251 (2015).
Smith, C. L. et al. Migration phenotype of brain-cancer cells predicts patient outcomes. Cell Rep. 15, 2616–2624 (2016).
Yankaskas, C. L. et al. A microfluidic assay for the quantification of the metastatic propensity of breast cancer specimens. Nat. Biomed. Eng. 3, 452–465 (2019).
Paul, C. D. et al. Interplay of the physical microenvironment, contact guidance and intracellular signaling in cell decision making. FASEB J. 30, 2161–2170 (2016).
Wolf, K. et al. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 20, 931–941 (2009).
Jucker, M., Tian, M. & Ingram, D. K. Laminins in the adult and aged brain. Mol. Chem. Neuropathol. 28, 209–218 (1996).
Weigelin, B., Bakker, G. J. & Friedl, P. Intravital third harmonic generation microscopy of collective melanoma cell invasion: principles of interface guidance and microvesicle dynamics. Intravital 1, 32–43 (2012).
Xie, Q., Mittal, S. & Berens, M. E. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 16, 1575–1584 (2014).
Inwald, E. C. et al. Ki-67 is a prognostic parameter in breast cancer patients: results of a large population-based cohort of a cancer registry. Breast Cancer Res. Treat. 139, 539–552 (2013).
Zhu, P., Du, X. L., Lu, G. & Zhu, J. J. Survival benefit of glioblastoma patients after FDA approval of temozolomide concomitant with radiation and bevacizumab: a population-based study. Oncotarget 8, 44015–44031 (2017).
Steeg, P. S. Targeting metastasis. Nat. Rev. Cancer 16, 201–218 (2016).
Paul, C. D., Mistriotis, P. & Konstantopoulos, K. Cancer cell motility: lessons from migration in confined spaces. Nat. Rev. Cancer 17, 131–140 (2017).
Liu, J. C., Zacksenhouse, M., Eisen, A., Nofech-Mozes, S. & Zacksenhaus, E. Identification of cell proliferation, immune response and cell migration as critical pathways in a prognostic signature for HER2+:ERα− breast cancer. PLoS ONE 12, e0179 (2017).
Shah, S. R. et al. 217 YAP is ready to Rac and Rho: elucidation of a novel YAP-driven network that potentiates brain cancer cell dispersal and confers poor survival in patients. Neurosurgery 63, 185–185 (2016).
Armento, A., Ehlers, J., Schotterl, S. & Naumann, U. in Glioblastoma (ed. De Vleeschouwer, S.) (Codon Publications, 2017).
Gritsenko, P., Leenders, W. & Friedl, P. Recapitulating in vivo-like plasticity of glioma cell invasion along blood vessels and in astrocyte-rich stroma. Histochem. Cell Biol. 148, 395–406 (2017).
Friedl, P. & Alexander, S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147, 992–1009 (2011).
Friedlander, D. R. et al. Migration of brain tumor cells on extracellular matrix proteins in vitro correlates with tumor type and grade and involves alphaV and beta1 integrins. Cancer Res. 56, 1939–1947 (1996).
Wong, E. et al. Cut-point for Ki-67 proliferation index as a prognostic marker for glioblastoma. Asia Pac. J. Clin. Oncol. 15, 5–9 (2019).
Abubakar, M. et al. Prognostic value of automated KI67 scoring in breast cancer: a centralised evaluation of 8,088 patients from 10 study groups. Breast Cancer Res. 18, 104 (2016).
Lin, J. G. et al. Linking invasive motility to protein expression in single tumor cells. Lab Chip 18, 371–384 (2018).
Shah, S. R. et al. Brachyury-YAP regulatory axis drives stemness and growth in cancer. Cell Rep. 21, 495–507 (2017).
Mistriotis, P. et al. Confinement hinders motility by inducing RhoA-mediated nuclear influx, volume expansion and blebbing. J. Cell Biol. 218, 4093–4111 (2019).
Tong, Z. et al. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS ONE 7, e29211 (2012).
Zhao, R. et al. Cell sensing and decision-making in confinement: the role of TRPM7 in a tug of war between hydraulic pressure and cross-sectional area. Sci. Adv. 5, eaaw7243 (2019).
Chen, S. H., Hung, W. C., Wang, P., Paul, C. & Konstantopoulos, K. Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Sci. Rep. 3, 1870 (2013).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009).
Eden, E., Lipson, D., Yogev, S. & Yakhini, Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 3, e39 (2007).
Acknowledgements
This line of research was supported by the National Cancer Institute through grant nos. R01-CA216855 (to A.Q.-H. and K.K.), R01-CA183804 (to K.K.) and R01-CA124704 (to S.S.M.) and Department of Defense grant no. CA160997 (to V.K.B.). A.Q.-H. was also supported by the Mayo Clinic Clinician Investigator Award and the William J. and Charles H. Mayo Professorship.
Author information
Authors and Affiliations
Contributions
B.S.W., S.R.S., A.Q.-H. and K.K. designed the study. B.S.W. performed most experiments, interpreted the data and wrote the manuscript. S.R.S. performed select experiments, conducted patient clinical analysis and assisted in writing the manuscript. C.L.Y. performed experiments using MAqCI, the transwell assay and isolation of cells for RNA-seq, and contributed to data analysis. D.C. performed select experiments. P.-H.W. and B.I. performed survival analyses based on RNA-seq data. K.R. and P.S. contributed to the design of the study and collected patient data. S.S.M. designed and provided support for the transwell-migration assay, while V.K.B., C.-M.F. and X.Z. performed the RNA-seq experiments and analysis. All authors interpreted data, provided critical insights and edited the manuscript. K.K. supervised the study and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
MAqCI is the subject of US utility patent applications 15/780,768 and 14/906,055. Intellectual property related to MAqCI is owned by the Johns Hopkins University and licenced to RecurX Bio, Inc., of which K.K. is a co-founder, consultant and board member. K.K. has a financial interest in RecurX Bio, Inc., which is subject to certain restrictions under university policy. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures, tables and video captions.
Supplementary Dataset 1
Measures of performance, patient characteristics and list of differentially expressed genes from two patients.
Supplementary Video 1
Migration of patient-derived primary GBM cells (GBM714) in MAqCI.
Rights and permissions
About this article
Cite this article
Wong, B.S., Shah, S.R., Yankaskas, C.L. et al. A microfluidic cell-migration assay for the prediction of progression-free survival and recurrence time of patients with glioblastoma. Nat Biomed Eng 5, 26–40 (2021). https://doi.org/10.1038/s41551-020-00621-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-020-00621-9
This article is cited by
-
Development of preoperative and postoperative models to predict recurrence in postoperative glioma patients: a longitudinal cohort study
BMC Cancer (2024)
-
Analytical device miniaturization for the detection of circulating biomarkers
Nature Reviews Bioengineering (2023)
-
Micro-engineering and nano-engineering approaches to investigate tumour ecosystems
Nature Reviews Cancer (2023)
-
Cells in the polyaneuploid cancer cell (PACC) state have increased metastatic potential
Clinical & Experimental Metastasis (2023)
-
Perivascular invasion of primary human glioblastoma cells in organotypic human brain slices: human cells migrating in human brain
Journal of Neuro-Oncology (2023)