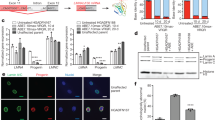
Abstract
The success of base editors for the study and treatment of genetic diseases depends on the ability to deliver them in vivo to the relevant cell types. Delivery via adeno-associated viruses (AAVs) is limited by AAV packaging capacity, which precludes the use of full-length base editors. Here, we report the application of dual AAVs for the delivery of split cytosine and adenine base editors that are then reconstituted by trans-splicing inteins. Optimized dual AAVs enable in vivo base editing at therapeutically relevant efficiencies and dosages in the mouse brain (up to 59% of unsorted cortical tissue), liver (38%), retina (38%), heart (20%) and skeletal muscle (9%). We also show that base editing corrects, in mouse brain tissue, a mutation that causes Niemann–Pick disease type C (a neurodegenerative ataxia), slowing down neurodegeneration and increasing lifespan. The optimized delivery vectors should facilitate the efficient introduction of targeted point mutations into multiple tissues of therapeutic interest.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the results of this study are available within the paper and its Supplementary Information. All unmodified reads for sequencing-based data in the manuscript are available from the NCBI Sequence Read Archive under accession number PRJNA532891. AAV genome sequences are provided in the Supplementary Information. Key plasmids from this work will be available from Addgene (depositor: D.R.L.), and other plasmids and raw data are available from the corresponding author on request.
Code availability
The custom code used in this study is provided in the Supplementary Information.
References
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846 (2018).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016).
Ryu, S. M. et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 36, 536–539 (2018).
Yeh, W. H., Chiang, H., Rees, H. A., Edge, A. S. B. & Liu, D. R. In vivo base editing of post-mitotic sensory cells. Nat. Commun. 9, 2184 (2018).
Chadwick, A. C., Wang, X. & Musunuru, K. In vivo base editing of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing. Arterioscl. Thromb. Vas. 37, 1741–1747 (2017).
Russell, S. et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860 (2017).
Carvalho, L. S. et al. Evaluating efficiencies of dual AAV approaches for retinal targeting. Front. Neurosci. 11, 503 (2017).
Wu, Z., Yang, H. & Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86 (2010).
Liu, D. R., Levy, J, M. & Yeh, W. H. AAV delivery of nucleobase editors. US patent 15/784,033 (2017).
Truong, D. J. J. et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 43, 6450–6458 (2015).
Zetsche, B., Volz, S. E. & Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142 (2015).
Wright, A. V. et al. Rational design of a split-Cas9 enzyme complex. Proc. Natl Acad. Sci. USA 112, 2984–2989 (2015).
Zettler, J., Schutz, V. & Mootz, H. D. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 583, 909–914 (2009).
Davis, K. M., Pattanayak, V., Thompson, D. B., Zuris, J. A. & Liu, D. R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 11, 316–318 (2015).
Stevens, A. J. et al. Design of a split intein with exceptional protein splicing activity. J. Am. Chem. Soc. 138, 2162–2165 (2016).
Shah, N. H., Eryilmaz, E., Cowburn, D. & Muir, T. W. Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. J. Am. Chem. Soc. 135, 5839–5847 (2013).
Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR–Cas9. Nat. Biotechnol. 33, 102–106 (2015).
Kim, Y. B. et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35, 371–376 (2017).
Villiger, L. et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 24, 1519–1525 (2018).
Grieger, J. C. & Samulski, R. J. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 79, 9933–9944 (2005).
Deverman, B. E. et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016).
Choi, J. H. et al. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol. Brain 7, 17 (2014).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Rees, H. A. et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790 (2017).
Gray, S. J. et al. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 22, 1143–1153 (2011).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Mendell, J. R. et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 377, 1713–1722 (2017).
Wu, Z., Asokan, A. & Samulski, R. J. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 14, 316–327 (2006).
Duan, D. Systemic AAV micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol. Ther. 26, 2337–2356 (2018).
Garvilles, R. G. et al. Dual functions of the RFTS domain of Dnmt1 in replication-coupled DNA methylation and in protection of the genome from aberrant methylation. PLoS ONE 10, e0137509 (2015).
Feng, J. et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430 (2010).
Inagaki, K. et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 14, 45–53 (2006).
Duan, D., Yue, Y. & Engelhardt, J. F. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 4, 383–391 (2001).
Xu, Z. et al. Trans-splicing adeno-associated viral vector-mediated gene therapy is limited by the accumulation of spliced mRNA but not by dual vector coinfection efficiency. Hum. Gene Ther. 15, 896–905 (2004).
Van Putten, M. et al. Low dystrophin levels increase survival and improve muscle pathology and function in dystrophin/utrophin double-knockout mice. FASEB J. 27, 2484–2495 (2013).
Li, D., Yue, Y. & Duan, D. Marginal level dystrophin expression improves clinical outcome in a strain of dystrophin/utrophin double knockout mice. PLoS ONE 5, e15286 (2010).
Tuchman, M., Jaleel, N., Morizono, H., Sheehy, L. & Lynch, M. G. Mutations and polymorphisms in the human ornithine transcarbamylase gene. Hum. Mutat. 19, 93–107 (2002).
Treacy, E. P. et al. Analysis of phenylalanine hydroxylase genotypes and hyperphenylalaninemia phenotypes using l-[1-13C]phenylalanine oxidation rates in vivo: a pilot study1. Pediatr. Res. 42, 430–435 (1997).
Hamman, K. et al. Low therapeutic threshold for hepatocyte replacement in murine phenylketonuria. Mol. Ther. 12, 337–344 (2005).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Asico, L. D. et al. Nephron segment-specific gene expression using AAV vectors. Biochem. Biophys. Res. Commun. 497, 19–24 (2018).
Foust, K. D. et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 27, 59–65 (2009).
Mercuri, E. et al. Nusinersen versus sham control in later-onset spinal muscular atrophy. N. Engl. J. Med. 378, 625–635 (2018).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Hordeaux, J. et al. The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 26, 664–668 (2018).
Huang, Q. et al. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. PLoS ONE 14, e0225206 (2019).
Harvey, R. J. & Napper, R. M. Quantitative study of granule and Purkinje cells in the cerebellar cortex of the rat. J. Comp. Neurol. 274, 151–157 (1988).
Vogel, M. W., Sunter, K. & Herrup, K. Numerical matching between granule and Purkinje cells in lurcher chimeric mice: a hypothesis for the trophic rescue of granule cells from target-related cell death. J. Neurosci. 9, 3454–3462 (1989).
Kim, J. Y. et al. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. Eur. J. Neurosci. 37, 1203–1220 (2013).
Kim, J. Y., Grunke, S. D., Levites, Y., Golde, T. E. & Jankowsky, J. L. Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. J. Vis. Exp. 91, 51863 (2014).
Hoxha, E., Balbo, I., Miniaci, M. C. & Tempia, F. Purkinje cell signaling deficits in animal models of ataxia. Front. Syn. Neurosci. 10, 6 (2018).
Matilla-Duenas, A. et al. Consensus paper: pathological mechanisms underlying neurodegeneration in spinocerebellar ataxias. Cerebellum 13, 269–302 (2014).
Chakrabarty, P. et al. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS ONE 8, e67680 (2013).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Zinn, E. et al. In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep. 12, 1056–1068 (2015).
Koch, S. F. et al. Genetic rescue models refute nonautonomous rod cell death in retinitis pigmentosa. Proc. Natl Acad. Sci. USA 114, 5259–5264 (2017).
Maeder, M. L. et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 25, 229–233 (2019).
Park, W. D. et al. Identification of 58 novel mutations in Niemann–Pick disease type C: correlation with biochemical phenotype and importance of PTC1-like domains in NPC1. Hum. Mutat. 22, 313–325 (2003).
Praggastis, M. et al. A murine Niemann–Pick C1 I1061T knock-in model recapitulates the pathological features of the most prevalent human disease allele. J. Neurosci. 35, 8091–8106 (2015).
Yu, T., Shakkottai, V. G., Chung, C. & Lieberman, A. P. Temporal and cell-specific deletion establishes that neuronal Npc1 deficiency is sufficient to mediate neurodegeneration. Hum. Mol. Genet. 20, 4440–4451 (2011).
Loftus, S. K. et al. Rescue of neurodegeneration in Niemann–Pick C mice by a prion-promoter-driven Npc1 cDNA transgene. Hum. Mol. Genet. 11, 3107–3114 (2002).
Lopez, M. E., Klein, A. D., Dimbil, U. J. & Scott, M. P. Anatomically defined neuron-based rescue of neurodegenerative Niemann–Pick type C disorder. J. Neurosci. 31, 4367–4378 (2011).
Elrick, M. J. et al. Conditional Niemann–Pick C mice demonstrate cell autonomous Purkinje cell neurodegeneration. Hum. Mol. Genet. 19, 837–847 (2010).
Ko, D. C. et al. Cell-autonomous death of cerebellar Purkinje neurons with autophagy in Niemann–Pick type C disease. PLoS Genet. 1, 81–95 (2005).
Langmade, S. J. et al. Pregnane X receptor (PXR) activation: a mechanism for neuroprotection in a mouse model of Niemann–Pick C disease. Proc. Natl Acad. Sci. USA 103, 13807–13812 (2006).
Hughes, M. P. et al. AAV9 intracerebroventricular gene therapy improves lifespan, locomotor function and pathology in a mouse model of Niemann–Pick type C1 disease. Hum. Mol. Genet. 27, 3079–3098 (2018).
Ling, C. et al. High-efficiency transduction of primary human hematopoietic stem/progenitor cells by AAV6 vectors: strategies for overcoming donor-variation and implications in genome editing. Sci. Rep. 6, 35495 (2016).
Nathwani, A. C. et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 371, 1994–2004 (2014).
Chandler, R. J. et al. Systemic AAV9 gene therapy improves the lifespan of mice with Niemann–Pick disease, type C1. Hum. Mol. Genet 26, 52–64 (2017).
Xie, C., Gong, X. M., Luo, J., Li, B. L. & Song, B. L. AAV9-NPC1 significantly ameliorates Purkinje cell death and behavioral abnormalities in mouse NPC disease. J. Lipid Res. 58, 512–518 (2017).
Hinderer, C. et al. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 29, 285–298 (2018).
Manno, C. S. et al. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 12, 342–347 (2006).
Habib, N. et al. Massively parallel single-nucleus RNA-Seq with DroNc-Seq. Nat. Methods 14, 955–958 (2017).
Li, P. et al. Allele-specific CRISPR–Cas9 genome editing of the single-base P23H mutation for rhodopsin-associated dominant retinitis pigmentosa. CRISPR J. 1, 55–64 (2018).
Sommer, C., Strähle, C., Köthe, U. & Hamprecht, F. A. Ilastik: Interactive learning and segmentation toolkit. In Eighth IEEE International Symposium on Biomedical Imaging 230–233 (ISBI, 2011).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Tsai, S. Q. et al. CIRCLE-Seq: a highly sensitive in vitro screen for genome-wide CRISPR–Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Ullman-Cullere, M. H. & Foltz, C. J. Body condition scoring: a rapid and accurate method for assessing health status in mice. Lab. Anim. Sci. 49, 319–323 (1999).
Foltz, C. & Ullman-Cullere, M. Guidelines for assessing the health and condition of mice. Lab Animal 28, 28–32 (1998).
Acknowledgements
This work was supported by the US National Institutes of Health (grant nos. UG3 TR002636, U01 AI142756, RM1 HG009490, R01 EB022376 and R35 GM118062), St. Jude Collaborative Research Consortium, DARPA (HR0011-17-2-0049), Ono Pharma Foundation, Bill and Melinda Gates Foundation and Howard Hughes Medical Institute. We thank the Harvard Center for Biological Imaging for infrastructure and support. We thank F. Zhang, B. Deverman and K. Chan for equipment access, guidance and helpful discussions, M. Doan for image analysis assistance and A. Hamidi for assistance with editing the manuscript.
Author information
Authors and Affiliations
Contributions
J.M.L. designed the research, constructed the plasmids, produced the AAV and performed the HEK cell, mouse systemic and CNS injection experiments. W.-H.Y. performed all of the CBE 3T3 experiments, image analysis and off-target analysis. J.R.D. performed the ABE 3T3 experiments. L.W.K. constructed the plasmids and performed the HEK cell experiments. N.P., R.B. and E.H. performed the retinal experiments. J.C. conceived of the retinal experiments and performed the data analysis. Q.L. conceived of and performed the sub-retinal injection experiments and data analysis. D.R.L. designed and supervised the research. J.M.L., W.-H.Y. and D.R.L. wrote the manuscript. All authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.R.L. is a consultant and co-founder of Beam Therapeutics, Prime Medicine, Editas Medicine and Pairwise Plants, all of which are companies that use genome editing. D.R.L., J.M.L., W.-H.Y. and L.W.K. have filed patent applications on AAV systems for base editor delivery. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11, Supplementary Tables 1 and 2, Supplementary Methods, Supplementary Code and Supplementary Notes.
Rights and permissions
About this article
Cite this article
Levy, J.M., Yeh, WH., Pendse, N. et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat Biomed Eng 4, 97–110 (2020). https://doi.org/10.1038/s41551-019-0501-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-019-0501-5
This article is cited by
-
Eukaryotic-driven directed evolution of Cas9 nucleases
Genome Biology (2024)
-
Adeno-associated virus as a delivery vector for gene therapy of human diseases
Signal Transduction and Targeted Therapy (2024)
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
Continuous directed evolution of a compact CjCas9 variant with broad PAM compatibility
Nature Chemical Biology (2024)
-
Efficient prime editing in mouse brain, liver and heart with dual AAVs
Nature Biotechnology (2024)