Abstract
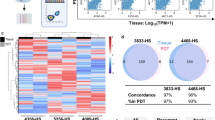
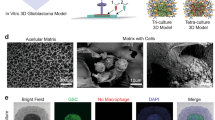
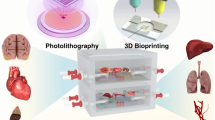
Patient-specific ex vivo models of human tumours that recapitulate the pathological characteristics and complex ecology of native tumours could help determine the most appropriate cancer treatment for individual patients. Here, we show that bioprinted reconstituted glioblastoma tumours consisting of patient-derived tumour cells, vascular endothelial cells and decellularized extracellular matrix from brain tissue in a compartmentalized cancer–stroma concentric-ring structure that sustains a radial oxygen gradient, recapitulate the structural, biochemical and biophysical properties of the native tumours. We also show that the glioblastoma-on-a-chip reproduces clinically observed patient-specific resistances to treatment with concurrent chemoradiation and temozolomide, and that the model can be used to determine drug combinations associated with superior tumour killing. The patient-specific tumour-on-a-chip model might be useful for the identification of effective treatments for glioblastoma patients resistant to the standard first-line treatment.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Code availability
The computer code for the bioprinting of the GBM-on-a-chip is provided as Supplementary Information.
Data availability
The authors declare that all data supporting the results in this study are available within the paper and its Supplementary information. The source data for the figures in this study are available from figshare (identifier https://doi.org/10.6084/m9.figshare.7392677)51.
References
Schreiber, S. L. et al. Towards patient-based cancer therapeutics. Nat. Biotechnol. 28, 904–906 (2010).
Al-Lazikani, B., Banerji, U. & Workman, P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 30, 679–692 (2012).
Creixell, P., Schoof, E. M., Erler, J. T. & Linding, R. Navigating cancer network attractors for tumor-specific therapy. Nat. Biotechnol. 30, 842–848 (2012).
Aparicio, S., Hidalgo, M. & Kung, A. L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer 15, 311–316 (2015).
Eirew, P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015).
Byrne, A. T. et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 17, 254–268 (2017).
Crystal, A. S. et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346, 1480–1486 (2014).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Hanahan, D. & Coussens, L. M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012).
Sackmann, E. K., Fulton, A. L. & Beebe, D. J. The present and future role of microfluidics in biomedical research. Nature 507, 181–189 (2014).
Picollet-D’hahan, N. et al. A 3D toolbox to enhance physiological relevance of human tissue models. Trends Biotechnol. 34, 757–769 (2016).
Garber, K. Between disease and a dish. Nat. Biotechnol. 32, 712–715 (2014).
Yamada, K. M. & Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 130, 601–610 (2007).
Lathia, J. D., Heddleston, J. M., Venere, M. & Rich, J. N. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell 8, 482–485 (2011).
Junttila, M. R. & de Sauvage, F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013).
Korolev, K. S., Xavier, J. B. & Gore, J. Turning ecology and evolution against cancer. Nat. Rev. Cancer 14, 371–380 (2014).
Ananthanarayanan, B., Kim, Y. & Kumar, S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials 32, 7913–7923 (2011).
Kievit, F. M. et al. Chitosan–alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials 31, 5903–5910 (2010).
Lee, G. Y., Kenny, P. A., Lee, E. H. & Bissell, M. J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4, 359–365 (2007).
Pedron, S., Becka, E. & Harley, B. A. Spatially gradated hydrogel platform as a 3D engineered tumor microenvironment. Adv. Mater. 27, 1567–1572 (2015).
Bhatia, S. N. & Ingber, D. E. Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772 (2014).
Shin, Y. et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 7, 1247–1259 (2012).
Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 18, v1–v75 (2016).
Ott, H. C. et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat. Med. 14, 213–221 (2008).
Chen, H. J. et al. A recellularized human colon model identifies cancer driver genes. Nat. Biotechnol. 34, 845–851 (2016).
Dunne, L. W. et al. Human decellularized adipose tissue scaffold as a model for breast cancer cell growth and drug treatments. Biomaterials 35, 4940–4949 (2014).
Pati, F. et al. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat. Commun. 5, 3935 (2014).
Choi, Y.-J. et al. 3D cell printing of functional skeletal muscle constructs using skeletal muscle-derived bioink. Adv. Healthc. Mater. 5, 2636–2645 (2016).
Le, P. U. et al. DRR drives brain cancer invasion by regulating cytoskeletal-focal adhesion dynamics. Oncogene 29, 4636–4647 (2010).
Jung, J. W., Lee, J.-S. & Cho, D.-W. Computer-aided multiple-head 3D printing system for printing of heterogeneous organ/tissue constructs. Sci. Rep. 6, 21685 (2016).
Kim, D. et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature 520, 363–367 (2015).
Jain, R. K. et al. Angiogenesis in brain tumours. Nat. Rev. Neurosci. 8, 610–622 (2007).
Brandes, A. A. et al. First-line chemotherapy with cisplatin plus fractionated temozolomide in recurrent glioblastoma multiforme: a phase II study of the Gruppo Italiano Cooperativo di Neuro-oncologia. J. Clin. Oncol. 22, 1598–1604 (2004).
Bao, Q. et al. Recombinant TIMP-1-GPI inhibits growth of fibrosarcoma and enhances tumor sensitivity to doxorubicin. Target. Oncol. 9, 251–261 (2014).
McFaline-Figueroa, J. L. et al. Minor changes in expression of the mismatch repair protein MSH2 exert a major impact on glioblastoma response to temozolomide. Cancer Res. 75, 3127–3138 (2015).
Munoz, J. L. et al. Temozolomide resistance in glioblastoma cells occurs partly through epidermal growth factor receptor-mediated induction of connexin 43. Cell Death Dis. 5, e1145 (2014).
Perazzoli, G. et al. Temozolomide resistance in glioblastoma cell lines: implication of MGMT, MMR, P-glycoprotein and CD133 expression. PLoS ONE 10, e0140131 (2015).
Hirose, Y., Berger, M. S. & Pieper, R. O. Abrogation of the Chk1-mediated G2 checkpoint pathway potentiates temozolomide-induced toxicity in a p53-independent manner in human glioblastoma cells. Cancer Res. 61, 5843–5849 (2001).
Vecchio, D. et al. Predictability, efficacy and safety of radiosensitization of glioblastoma-initiating cells by the ATM inhibitor KU-60019. Int. J. Cancer 135, 479–491 (2014).
Hegi, M. E. et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin. Cancer Res. 10, 1871–1874 (2004).
Eads, J. R. et al. Phase I clinical trial of temozolomide and methoxyamine (TRC-102) in patients with advanced solid tumors. J. Clin. Oncol. 33, 2558–2558 (2015).
Xu, Z. et al. Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials 34, 4109–4117 (2013).
Yu, M. et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014).
Choi, Y.-J., Yi, H.-G., Kim, S.-W. & Cho, D.-W. 3D cell printed tissue analogues: a new platform for theranostics. Theranostics 7, 3118–3137 (2017).
Lind, J. U. et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater. 16, 303–308 (2016).
Lee, H. & Cho, D.-W. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip 16, 2618–2625 (2016).
Kang, T.-Y., Hong, J. M., Jung, J. W., Yoo, J. J. & Cho, D.-W. Design and assessment of a microfluidic network system for oxygen transport in engineered tissue. Langmuir 29, 701–709 (2013).
Jung, J. W. et al. Evaluation of the effective diffusivity of a freeform fabricated scaffold using computational simulation. J. Biomech. Eng. 135, 084501 (2013).
Kim, J. Y. & Cho, D.-W. Blended PCL/PLGA scaffold fabrication using multi-head deposition system. Microelectron. Eng. 86, 1447–1450 (2009).
Jo, H. Y. et al. The unreliability of MTT assay in the cytotoxic test of primary cultured glioblastoma cells. Exp. Neurobiol. 24, 235–245 (2015).
Yi, H.-G. et al. A bioprinted human-glioblastoma-on-a-chip reproduces patient-specific responses to chemoradiotherapy. Figshare https://doi.org/10.6084/m9.figshare.7392677 (2018).
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government, MSIP (grant nos 2010-0018294, 2015R1A2A2A01005515 and 2018R1A2B2009540). This study was partly supported by the Technology Innovation Program (grant no. 10050154, Business Model Development for Personalized Medicine Based on Integrated Genome and Clinical Information) and by the Bio and Medical Technology Development Program of the NRF funded by the Korean government, MSIP (grant no. 2015M3C7A1028926). We thank J. M. Hong for technical assistance and M. N. Park for helpful discussions.
Author information
Authors and Affiliations
Contributions
H.-G.Y., D.-W.C. and S.H.Paek conceived the concept of applying 3D-printing technology to establish the patient-specific GBM-on-a-chip. H.-G.Y. and Y.H.J. devised the working principles of the chip in detail. H.-G.Y. designed and performed most of the experiments. Y.K. performed the bioinformatics analyses and wrote the relevant results and methods. Y.-J.C. assisted with the characterization of the BdECM bioink, 3D printing of the GBMs-on-chips and performed the tumour spheroid invasion study. H.E.M. prepared for the IRB approval process to conduct the experiments using patient-derived GBM cells and organized the clinical information of the patients. S.H.Paek was the physician in charge of the GBM patients. S.H.Park performed the pathological detection and analysis of the patient-derived GBMs. K.S.K. contributed to the discussion for the initial stages of this work. M.B. assisted with the 3D printing and culturing of the GBMs-on-chips. J.J. contributed to the discussion for the revisions of the manuscript. H.Y. provided the genetic analysis data of patient-derived GBM cells. H.-G.Y., Y.H.J., S.H.Paek and D.-W.C. analysed the data. S.H.Paek also analysed the clinical observations and provided the relevant consultation. D.-W.C. provided overall guidance and supervised the project. H.-G.Y. and Y.H.J. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Patents on the use of BdECM bioink in modelling cancer (patent no. 10-1860798, Korea) and on 3D printing of GBM-on-a-chip (patent no. 10-1803618, Korea) have been registered.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures, tables, methods and code.
Supplementary Video 1
Cell-printing of a glioblastoma-on-a-chip.
Supplementary Video 2
Migration of glioblastoma cells to the surrounding matrix.
Rights and permissions
About this article
Cite this article
Yi, HG., Jeong, Y.H., Kim, Y. et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng 3, 509–519 (2019). https://doi.org/10.1038/s41551-019-0363-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-019-0363-x
This article is cited by
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
3D bioprinted tumor model: a prompt and convenient platform for overcoming immunotherapy resistance by recapitulating the tumor microenvironment
Cellular Oncology (2024)
-
The trend of allogeneic tendon decellularization: literature review
Cell and Tissue Banking (2024)
-
Micro-engineering and nano-engineering approaches to investigate tumour ecosystems
Nature Reviews Cancer (2023)
-
Controlled tumor heterogeneity in a co-culture system by 3D bio-printed tumor-on-chip model
Scientific Reports (2023)