Abstract
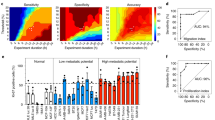
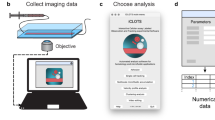
The risk stratification of prostate cancer and breast cancer tumours from patients relies on histopathology, selective genomic testing, or on other methods employing fixed formalin tissue samples. However, static biomarker measurements from bulk fixed-tissue samples provide limited accuracy and actionability. Here, we report the development of a live-primary-cell phenotypic-biomarker assay with single-cell resolution, and its validation with prostate cancer and breast cancer tissue samples for the prediction of post-surgical adverse pathology. The assay includes a collagen-I/fibronectin extracellular-matrix formulation, dynamic live-cell biomarkers, a microfluidic device, machine-vision analysis and machine-learning algorithms, and generates predictive scores of adverse pathology at the time of surgery. Predictive scores for the risk stratification of 59 prostate cancer patients and 47 breast cancer patients, with values for area under the curve in receiver-operating-characteristic curves surpassing 80%, support the validation of the assay and its potential clinical applicability for the risk stratification of cancer patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. Anonymized biomarker quantifications are available upon request from the corresponding author.
References
Siegel, R., Ma, J., Zou, Z. & Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 (2014).
Bell, N.et al. Recommendations on screening for prostate cancer with the prostate-specific antigen test. CMAJ 186, 1225–1234 2014).
Mariotto, A. B. et al. Cancer survival: an overview of measures, uses, and interpretation. J. Natl Cancer Inst. Monogr. 2014, 145–186 (2014).
Shieh, Y. et al. Population-based screening for cancer: hope and hype. Nat. Rev. Clin. Oncol. 13, 550–565 (2016).
U.S. Cancer Statistics Working Group United States Cancer Statistics: 1999–2013 Incidence and Mortality Web-based Report http://www.cdc.gov/uscs (U.S. Department of Health and Human Services, 2016).
Canfield, S. E. et al. A guide for clinicians in the evaluation of emerging molecular diagnostics for newly diagnosed prostate cancer. Rev. Urol. 16, 172–180 (2014).
Dahabreh, I. J. et al. Core Needle and Open Surgical Biopsy for Diagnosis of Breast Lesions: An Update to the 2009 Report AHRQ Comparative Effectiveness Reviews (Agency for Healthcare Research and Quality, 2014).
Alvarado, M., Ozanne, E. & Esserman, L. Overdiagnosis and overtreatment of breast cancer. Am. Soc. Clin. Oncol. Educ. Book 2012, e40–e45 (2012).
Evans, A. & Vinnicombe, S. Overdiagnosis in breast imaging. Breast 31, 270–273 (2017).
Hugosson, J. & Carlsson, S. Overdetection in screening for prostate cancer. Curr. Opin. Urol. 24, 256–263 (2014).
Klotz, L. Cancer overdiagnosis and overtreatment. Curr. Opin. Urol. 22, 203–209 (2012).
Lebeau, A. & Kuhn, T. Updates in the treatment of ductal carcinoma in situ of the breast. Curr. Opin. Obstet. Gynecol. 28, 49–58 (2016).
Strope, S. A. & Andriole, G. L. Prostate cancer screening: current status and future perspectives. Nat. Rev. Urol. 7, 487–493 (2010).
Verma, M., Patel, P. & Verma, M. Biomarkers in prostate cancer epidemiology. Cancers 3, 3773–3798 (2011).
Koren, S. & Bentires-Alj, M. Breast tumor heterogeneity: source of fitness, hurdle for therapy. Mol. Cell 60, 537–546 (2015).
Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334 (2012).
Zbuk, K. M. & Eng, C. Cancer phenomics: RET and PTEN as illustrative models. Nat. Rev. Cancer 7, 35–45 (2007).
Albala, D., Knopf, K. & Sant, G. Phenotypic cancer biomarkers-future role in precision oncology?. NPJ Precision Oncology 1, 21 (2017).
Chander, A. et al. Rapid and short-term extra-cellular matrix-mediated in vitro culturing of tumor and non-tumor human primary prostate cells from fresh radical prostatectomy tissue. Urology 105, 91–100 (2017).
Chander, A. C. et al. Systems, devices and methods for microfluidic culturing, manipulation and analysis of tissues and cells. US patent US20130149724A1 (2013).
Chander, A. C. et al. Systems, devices and methods for microfluidic culturing, manipulation and analysis of tissues and cells. US patent US20160272934A1 (2014).
Chander, A. C. Systems, methods and devices for measuring growth/oncogenic and migration/metastatic potential. US patent US20130237453A1 (2011).
Chander, A. C. Integrin-Linked Kinase, ECM Compostion and Substrate Rigidity Regulate Focal Adhesion—Actin Coupling, Modulating Survival, Proliferation andMigration: Towards a Biophysical Cancer Biomarker. PhD thesis, Columbia Univ. (2012).
Abdeen, A. A., Lee, J. & Kilian, K. A. Capturing extracellular matrix properties in vitro: microengineering materials to decipher cell and tissue level processes. Exp. Biol. Med. 241, 930–938 (2016).
Kacsinta, A. D. et al. Intracellular modifiers of integrin alpha 6p production in aggressive prostate and breast cancer cell lines. Biochem. Biophys. Res. Commun. 454, 335–340 (2014).
Chander, A. C., Su, W. R. & Varsanik, J. S. Cell imaging and analysis to differentiate clinically relevant sub-populations of cells. Patent WO2016138041A2 (2015).
Cuzick, J. et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 12, 245–255 (2011).
Klein, E. A. et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol. 66, 550–560 (2014).
Little, J. et al. Multigene panels in prostate cancer risk assessment: a systematic review. Genet. Med. 18, 535–544 (2016).
Mendhiratta, N., Meng, X. & Taneja, S. S. Using multiparametric MRI to ‘personalize’ biopsy for men. Curr. Opin. Urol. 25, 498–503 (2015).
Ross, A. E. et al. A genomic classifier predicting metastatic disease progression in men with biochemical recurrence after prostatectomy. Prostate Cancer Prostatic. Dis. 17, 64–69 (2014).
Scarpato, K. R. & Barocas, D. A. Use of mpMRI in active surveillance for localized prostate cancer. Urol. Oncol. 34, 320–325 (2016).
Sternberg, I. A., Vela, I. & Scardino, P. T. Molecular profiles of prostate cancer: to treat or not to treat. Annu. Rev. Med. 67, 119–135 (2016).
Zhuang, L. & Johnson, M. T. How precisely can prostate cancer be managed? Int. Neurourol. J. 20, S120–S130 (2016).
Wong, L. M. et al. Evaluation of models predicting insignificant prostate cancer to select men for active surveillance of prostate cancer. Prostate Cancer Prostatic Dis. 18, 137–143 (2015).
Tosoian, J. J., Carter, H. B., Lepor, A. & Loeb, S. Active surveillance for prostate cancer: current evidence and contemporary state of practice. Nat. Rev. Urol. 13, 205–215 (2016).
Street, C. A. & Bryan, B. A. Rho kinase proteins—pleiotropic modulators of cell survival and apoptosis. Anticancer Res. 31, 3645–3657 (2011).
Wirtz, D., Konstantopoulos, K. & Searson, P. C. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 11, 512–522 (2011).
Kang, J. et al. Improving drug discovery with high-content phenotypic screens by systematic selection of reporter cell lines. Nat. Biotechnol. 34, 70–77 (2016).
Almendro, V., Marusyk, A. & Polyak, K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 8, 277–302 (2013).
Cross, S. E., Jin, Y. S., Rao, J. & Gimzewski, J. K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotech. 2, 780–783 (2007).
Akobeng, A. K. Understanding diagnostic tests 1: sensitivity, specificity and predictive values. Acta Paediatr. 96, 338–341 (2007).
Akobeng, A. K. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr. 96, 644–647 (2007).
Akobeng, A. K. Understanding diagnostic tests 2: likelihood ratios, pre- and post-test probabilities and their use in clinical practice. Acta Paediatr. 96, 487–491 (2007).
Liu, A. Y. & True, L. D. Characterization of prostate cell types by CD cell surface molecules. Am. J. Pathol. 160, 37–43 (2002).
Signoretti, S. & Loda, M. Defining cell lineages in the prostate epithelium. Cell Cycle 5, 138–141 (2006).
Criminisi, A. & Shotton, J. Decision Forests for Computer Vision and Medical Image Analysis (Springer, New York, NY, 2013).
Ho, T. K. Proceedings of 3rd International Conference on Document Analysis and Recognition Vol. 2, A1–A7 (IEEE, Montreal, 1995)..
Ho, T. K. The random subspace method for constructing decision forests. IEEE. Trans. Pattern. Anal. Mach. Intell. 20, 832–844 (1998).
Acknowledgements
Tissue samples were provided by Lahey Hospital and Medical Center, Department of Cancer Research, American Medical Professionals of New York, by Urology Place of San Antonio and by the NCI Cooperative Human Tissue Network (CHTN). The authors thank S.K. Sia for a thoughtful review of the manuscript and P. Chaturvedi, R. Gottileb and M.B. Lisman for discussions, as well as M. Foroohar, S. Zappala, H. Rashid, V. Mouraviev, K. Christ, T.B. Sullivan and N. Kella. M.S.M. thanks J.A. Manak, C.A. Manak, G.H. Manak, P.L. Manak, L.L. Manak and P.W. Manak for support. J.S.V. thanks S.H. Varsanik for support. A.C.C. thanks C.S. Chandersekaran, A.C. Chandersekaran, A.B. Cravens Chander and I.E.A. Chander for support.
Author information
Authors and Affiliations
Contributions
M.S.M. and J.S.V. carried out technology development, experimental work and data analysis. B.J.H., M.J.W. and W.R.S. performed technology development and experimental work. N.J., R.J.S., G.D. and T.M. carried out experimental work and data analysis. N.S. acquired samples and performed data analysis. A.M. and D.B. carried out sample acquisition. H.M.C. conducted data analysis and contributed to writing the manuscript. K.B.K. and D.M.A. provided clinical guidance and contributed to writing the manuscript. G.R.S. provided clinical guidance, data analysis and contributed to writing the manuscript. A.C.C. provided technology conception, technology development and project planning, and carried out experimental work and data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing financial interests: M.S.M., J.S.V., M.J.W., W.R.S., N.J., N.S., A.M., D.B., R.J.S., G.D., T.M., H.M.C., K.B.K., G.R.S. and A.C.C.: Cellanyx Diagnostics, stock options. B.J.H.: Cellanyx Diagnostics, consultant stock options. D.M.A.: Genomic Health, speaker; Myriad Genetics, speaker; Cellanyx Diagnostics, stock options; Applied Medical, stock options. The following patents WO2016138041, WO2013075145 and WO201204826921,22,26 cover aspects of the technology presented in this paper.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, figures, tables and references.
Rights and permissions
About this article
Cite this article
Manak, M.S., Varsanik, J.S., Hogan, B.J. et al. Live-cell phenotypic-biomarker microfluidic assay for the risk stratification of cancer patients via machine learning. Nat Biomed Eng 2, 761–772 (2018). https://doi.org/10.1038/s41551-018-0285-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-018-0285-z
This article is cited by
-
Diagnosing glaucoma in primary eye care and the role of Artificial Intelligence applications for reducing the prevalence of undetected glaucoma in Australia
Eye (2024)
-
Smartphone-based platforms implementing microfluidic detection with image-based artificial intelligence
Nature Communications (2023)
-
Towards the differential diagnosis of prostate cancer by the pre-treatment of human urine using ionic liquids
Scientific Reports (2020)
-
Artificial intelligence in healthcare
Nature Biomedical Engineering (2018)