Abstract
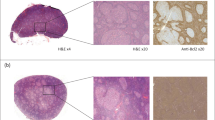
The identification of patients with aggressive cancer who require immediate therapy is a health challenge in low- and middle-income countries. Limited pathology resources, high healthcare costs and large caseloads call for the development of advanced stand-alone diagnostics. Here, we report and validate an automated, low-cost point-of-care device for the molecular diagnosis of aggressive lymphomas. The device uses contrast-enhanced microholography and a deep learning algorithm to directly analyse percutaneously obtained fine-needle aspirates. We show the feasibility and high accuracy of the device in cells, as well as the prospective validation of the results in 40 patients clinically referred for image-guided aspiration of nodal mass lesions suspicious of lymphoma. Automated analysis of human samples with the portable device should allow for the accurate classification of patients with benign and malignant adenopathy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hsu, C. Y., Jung, S. M. & Chuang, S. S. Physician supply and demand in anatomical pathology in Taiwan. J. Formos. Med. Assoc. 110, 78–84 (2011).
Nelson, A. M., Milner, D. A., Rebbeck, T. R. & Iliyasu, Y. Oncologic care and pathology resources in Africa: survey and recommendations. J. Clin. Oncol. 34, 20–26 (2016).
Varmus, H. & Kumar, H. S. Addressing the growing international challenge of cancer: a multinational perspective. Sci. Transl. Med. 5, 175cm2 (2013).
Livingston, J. Cancer in the shadow of the AIDS epidemic in southern Africa. Oncologist 18, 783–786 (2013).
Chabner, B., Dryden-Petersen, S. & Efstathiou, J. Cancer in Botswana: the second wave of AIDS in Sub-Saharan Africa. Oncologist 18, 777–778 (2013).
Naresh, K. N. et al. Lymphomas in sub-Saharan Africa—what can we learn and how can we help in improving diagnosis, managing patients and fostering translational research. Br. J. Haematol. 154, 696–703 (2011).
Carbone, A. et al. Diagnosis and management of lymphomas and other cancers in HIV-infected patients. Nat. Rev. Clin. Oncol. 11, 223–238 (2014).
Mwamba, P. M. et al. AIDS-related non-Hodgkin’s lymphoma in Sub-Saharan Africa: current status and realities of therapeutic approach. Lymphoma 2012, 904367 (2012).
D’Ambrosio, M. V. et al. Point-of-care quantification of blood-borne filarial parasites with a mobile phone microscope. Sci. Transl. Med. 7, 286re4 (2015).
Greenbaum, A. et al. Wide-field computational imaging of pathology slides using lens-free on-chip microscopy. Sci. Transl. Med. 6, 267ra175 (2014).
Im, H. et al. Digital diffraction analysis enables low-cost molecular diagnostics on a smartphone. Proc. Natl Acad. Sci. USA 112, 5613–5618 (2015).
Zheng, G., Lee, S. A., Antebi, Y., Elowitz, M. B. & Yang, C. The ePetri dish, an on-chip cell imaging platform based on subpixel perspective sweeping microscopy (SPSM). Proc. Natl Acad. Sci. USA 108, 16889–16894 (2011).
Tapley, A. et al. Mobile digital fluorescence microscopy for diagnosis of tuberculosis. J. Clin. Microbiol. 51, 1774–1778 (2013).
Laksanasopin, T. et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 7, 273re1 (2015).
Yeo, S. J. et al. Smartphone-based fluorescent diagnostic system for highly pathogenic H5N1 viruses. Theranostics 6, 231–242 (2016).
Kanakasabapathy, M. K. et al. An automated smartphone-based diagnostic assay for point-of-care semen analysis. Sci. Transl. Med. 9, eaai7863 (2017).
Priye, A. et al. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci. Rep. 7, 44778 (2017).
Kanakasabapathy, M. K. et al. Rapid, label-free CD4 testing using a smartphone compatible device. Lab Chip 17, 2910–2919 (2017).
Ming, K. et al. Integrated quantum dot barcode smartphone optical device for wireless multiplexed diagnosis of infected patients. ACS Nano 9, 3060–3074 (2015).
Ko, J. et al. Smartphone-enabled optofluidic exosome diagnostic for concussion recovery. Sci. Rep. 6, 31215 (2016).
Meda, B. A. et al. Diagnosis and subclassification of primary and recurrent lymphoma. The usefulness and limitations of combined fine-needle aspiration cytomorphology and flow cytometry. Am. J. Clin. Pathol. 113, 688–699 (2000).
Zeppa, P. et al. Fine needle aspiration cytology and flow cytometry immunophenotyping of non-Hodgkin lymphoma: can we do better? Cytopathology 21, 300–310 (2010).
Savage, E. C., Vanderheyden, A. D., Bell, A. M., Syrbu, S. I. & Jensen, C. S. Independent diagnostic accuracy of flow cytometry obtained from fine-needle aspirates: a 10-year experience with 451 cases. Am. J. Clin. Pathol. 135, 304–309 (2011).
Wei, Q. et al. Plasmonics enhanced smartphone fluorescence microscopy. Sci. Rep. 7, 2124 (2017).
Xu, W., Jericho, M. H., Meinertzhagen, I. A. & Kreuzer, H. J. Digital in-line holography for biological applications. Proc. Natl Acad. Sci. USA 98, 11301–11305 (2001).
Gurkan, U. A. et al. Miniaturized lensless imaging systems for cell and microorganism visualization in point-of-care testing. Biotechnol. J. 6, 138–149 (2011).
Greenbaum, A. et al. Imaging without lenses: achievements and remaining challenges of wide-field on-chip microscopy. Nat. Methods 9, 889–895 (2012).
Pathania, D. et al. Holographic assessment of lymphoma tissue (HALT) for global oncology field applications. Theranostics 6, 1603–1610 (2016).
Matasar, M. J. et al. Expert second-opinion pathology review of lymphoma in the era of the World Health Organization classification. Ann. Oncol. 23, 159–166 (2012).
Swerdllow, S. H. et al. (eds) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues 4th edn (IARC Press, Lyon, 2008).
Swerdlow, S. H. et al. (eds) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues Revised 4th edn (IARC Press, Lyon, 2017).
Demurtas, A. et al. Tissue flow cytometry immunophenotyping in the diagnosis and classification of non-Hodgkin’s lymphomas: a retrospective evaluation of 1,792 cases. Cytom. B 84, 82–95 (2013).
Van der Loos, C. M. Chromogens in multiple immunohistochemical staining used for visual assessment and spectral imaging: the colorful future. J. Histotechnol. 33, 31–40 (2010).
Brown, C. A. et al. Predictors of timely access of oncology services and advanced-stage cancer in an HIV-endemic setting. Oncologist 21, 731–738 (2016).
Eichenauer, D. A. et al. Hodgkin’s lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 25, iii70–iii75 (2014).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. In Proc. International Conference on Learning Representations (ICLR, 2015).
Acknowledgements
The authors thank J. Min for assay optimization, K. Joyes for editing the manuscript, and all members of MGH’s Division of Interventional Radiology, Department of Pathology and Cancer Center who contributed to patient care. This work was supported in part by 5UH2CA202637 (to R.W. and B.Chabner), 4R00CA201248 (to H.I.) and a grant from the V-Foundation for Cancer Research (to R.W. and C.M.C.). H.L. was supported in part by R21-CA205322, R01-HL113156 and the MGH scholar fund. A.K. has been supported by the Mac Erlaine Scholarship from the Academic Radiology Research Trust, St. Vincents Radiology Group, Dublin, Ireland, and also by the Higher Degree Bursary from the Faculty of Radiologists at the Royal College of Surgeons in Ireland.
Author information
Authors and Affiliations
Contributions
H.I., H.L., C.M.C. and R.W. designed the study. H.I., I.D. and B.Coble designed and fabricated the CEM device. D.P., P.J.M. and H.I. designed and optimized the experiments. H.I., D.P., P.J.M., S.H. and L.R. processed and analysed the samples. H.I., M.A., H.L., L.F. and M.P. established the computational algorithms. R.W., A.K., A.R.S., J.S.A., B.Chabner and C.M.C conducted the clinical trials. All authors reviewed the data. H.I. and R.W. wrote the paper, which was edited by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures and tables
Rights and permissions
About this article
Cite this article
Im, H., Pathania, D., McFarland, P.J. et al. Design and clinical validation of a point-of-care device for the diagnosis of lymphoma via contrast-enhanced microholography and machine learning. Nat Biomed Eng 2, 666–674 (2018). https://doi.org/10.1038/s41551-018-0265-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-018-0265-3
This article is cited by
-
Artificial intelligence performance in detecting lymphoma from medical imaging: a systematic review and meta-analysis
BMC Medical Informatics and Decision Making (2024)
-
Smartphone-based platforms implementing microfluidic detection with image-based artificial intelligence
Nature Communications (2023)
-
Multiplexed imaging in oncology
Nature Biomedical Engineering (2022)
-
The identification of gene signatures in patients with extranodal NK/T-cell lymphoma from a pair of twins
BMC Cancer (2021)
-
Machine learning for sperm selection
Nature Reviews Urology (2021)