Abstract
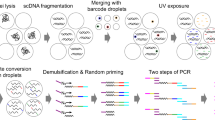
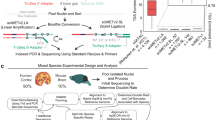
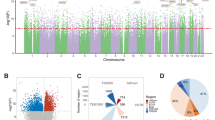
Methylomic analyses typically require substantial amounts of DNA, thus hindering studies involving scarce samples. Here, we show that microfluidic diffusion-based reduced representation bisulfite sequencing (MID-RRBS) permits high-quality methylomic profiling with nanogram-to-single-cell quantities of starting DNA. We used the microfluidic device, which allows for efficient bisulfite conversion with high DNA recovery, to analyse genome-wide DNA methylation in cell nuclei isolated from mouse brains and sorted into NeuN+ (primarily neuronal) and NeuN− (primarily glial) fractions, and to establish cell-type-specific methylomes. Genome-wide methylation and methylation in low-CpG-density promoter regions showed distinct patterns for NeuN+ and NeuN− fractions from the mouse cerebellum. The identification of substantial variations in the methylomic landscapes of the NeuN+ fraction of the frontal cortex of mice chronically treated with an atypical antipsychotic drug suggests that this technology can be broadly used for cell-type-specific drug profiling and for the study of drug–methylome interactions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hansen, K. D. et al. Increased methylation variation in epigenetic domains across cancer types. Nat. Genet. 43, 768–775 (2011).
Spiers, H. et al. Methylomic trajectories across human fetal brain development. Genome Res. 25, 338–352 (2015).
Numata, S. et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 90, 260–272 (2012).
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Lister, R. et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536 (2008).
Cokus, S. J. et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008).
Down, T. A. et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 26, 779–785 (2008).
Serre, D., Lee, B. H. & Ting, A. H. MBD-isolated genome sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 38, 391–399 (2010).
Meissner, A. et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33, 5868–5877 (2005).
Gu, H. et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods 7, 133–136 (2010).
Adey, A. & Shendure, J. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Res. 22, 1139–1143 (2012).
Guo, H., Zhu, P., Wu, X., Li, X., Wen, L. & Tang, F. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 23, 2126–2135 (2013).
Smallwood, S. A. et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014).
Farlik, M., Sheffield, Nathan, C., Nuzzo, A., Datlinger, P., Schönegger, A., Klughammer, J. & Bock, C. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 10, 1386–1397 (2015).
Luo, C. et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Lohr, J. G. et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 32, 479–484 (2014).
Han, L. et al. Co-detection and sequencing of genes and transcripts from the same single cells facilitated by a microfluidics platform. Sci. Rep. 4, 6485 (2014).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Gierahn, T. M. et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017).
Cao, Z., Chen, C., He, B., Tan, K. & Lu, C. A microfluidic device for epigenomic profiling using 100 cells. Nat. Methods 12, 959–962 (2015).
Rotem, A. et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 33, 1165–1172 (2015).
Yoon, J., Park, M. K., Lee, T. Y., Yoon, Y. J. & Shin, Y. LoMA-B: a simple and versatile lab-on-a-chip system based on single-channel bisulfite conversion for DNA methylation analysis. Lab Chip 15, 3530–3539 (2015).
Stark, A., Shin, D. J., Pisanic, T. II., Hsieh, K. & Wang, T. H. A parallelized microfluidic DNA bisulfite conversion module for streamlined methylation analysis. Biomed. Microdevices 18, 5 (2016).
Ehrich, M., Zoll, S., Sur, S. & van den Boom, D. A new method for accurate assessment of DNA quality after bisulfite treatment. Nucleic Acids Res. 35, e29 (2007).
Grunau, C., Clark, S. J. & Rosenthal, A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 29, e65 (2001).
Munson, K., Clark, J., Lamparska-Kupsik, K. & Smith, S. S. Recovery of bisulfite-converted genomic sequences in the methylation-sensitive qPCR. Nucleic Acids Res. 35, 2893–2903 (2007).
Ma, S. et al. Diffusion-based microfluidic PCR for “one-pot” analysis of cells. Lab Chip 14, 2905–2909 (2014).
Boyle, P. et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 13, R92 (2012).
Guo, S. et al. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 49, 635–642 (2017).
Zhou, J. et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 6, 10221 (2015).
Xu, R. H. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 16, 1155–1161 (2017).
Schillebeeckx, M. et al. Laser capture microdissection-reduced representation bisulfite sequencing (LCM-RRBS) maps changes in DNA methylation associated with gonadectomy-induced adrenocortical neoplasia in the mouse. Nucleic Acids Res. 41, e116 (2013).
Reizel, Y. et al. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes Dev. 29, 923–933 (2015).
Gabel, H. W. et al. Disruption of DNA methylation-dependent long gene repression in Rett syndrome. Nature 522, 89–93 (2015).
Mullen, R. J., Buck, C. R. & Smith, A. M. NeuN, a neuronal specific nuclear protein in vertebrates. Development 116, 201–211 (1992).
Weyer, A. & Schilling, K. Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J. Neurosci. Res. 73, 400–409 (2003).
Weber, M. et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39, 457–466 (2007).
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Davies, M. N. et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 13, R43 (2012).
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008).
Angermueller, C. et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016).
Meltzer, H. Y. Update on typical and atypical antipsychotic drugs. Annu. Rev. Med. 64, 393–406 (2013).
Kurita, M. et al. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat. Neurosci. 15, 1245–1254 (2012).
Kasowski, M. et al. Extensive variation in chromatin states across humans. Science 342, 750–752 (2013).
Feinberg, A. P. & Irizarry, R. A. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl Acad. Sci. USA 107, 1757–1764 (2010).
Oey, H., Isbel, L., Hickey, P., Ebaid, B. & Whitelaw, E. Genetic and epigenetic variation among inbred mouse littermates: identification of inter-individual differentially methylated regions. Epigenet. Chromat. 8, 54 (2015).
Millar, J. K. et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423 (2000).
Ayalew, M. et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol. Psychiatry 17, 887–905 (2012).
Cukier, H. N. et al. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol. Autism 5, 1 (2014).
Lee, B. J. et al. Analysis of differential gene expression mediated by clozapine in human postmortem brains. Schizophr. Res. 185, 58–66 (2017).
Unger, M. A., Chou, H. P., Thorsen, T., Scherer, A. & Quake, S. R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116 (2000).
Lake, B. B. et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Akalin, A. et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, 2012–2013 (2012).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Wu, H., Wang, C. & Wu, Z. A new shrinkage estimator for dispersion improves differential expression detection in RNA-seq data. Biostatistics 14, 232–243 (2013).
Dennis, G. et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, R60 (2003).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Samson, E., Marchand, J. & Snyder, K. A. Calculation of ionic diffusion coefficients on the basis of migration test results. Mater. Struct. 36, 156–165 (2003).
Hayduk, W. & Laudie, H. Prediction of diffusion coefficients for nonelectrolytes in dilute aqueous solutions. AIChE J. 20, 611–615 (1974).
Nkodo, A. E. et al. Diffusion coefficient of DNA molecules during free solution electrophoresis. Electrophoresis 22, 2424–2432 (2001).
Acknowledgements
We thank C. Luo, J. Lucero, M.M. Behrens and J.R. Ecker at Salk Institute and J. Ma at Carnegie Mellon University for helpful discussion. This work was supported by US National Institutes of Health grants EB017235 (C.L.), HG009256 (C.L.), MH084894 (J.G.-M.), MH111940 (J.G.-M.) and NS094574 (H.X.).
Author information
Authors and Affiliations
Contributions
C.L. and S.M. designed the microfluidic device. S.M. and C.L. developed the MID-RRBS protocol. C.L. supervised the research. C.L., S.M., J.G.-M. and H.X. designed the biological experiments. S.M. conducted experiments to generate MID-RRBS and mRNA-seq data on the cell/brain samples and performed data analysis. M.d.l.F.R. performed the clozapine treatment and sample collection. Z.S. helped with the device characterization. C.S. and T.W.M. helped with device fabrication and set-up. C.L. and S.M. wrote the manuscript. All authors proofread the manuscript and provided comments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures and tables.
Supplementary Table 5
List of genes associated with differentially CG-methylated regions (10 ng sample).
Supplementary Table 6
List of genes associated with differentially CG-methylated regions (0.5 ng sample).
Supplementary Table 7
GO term enrichment of genes associated with differentially CG methylated regions (10 ng sample).
Supplementary Table 8
GO term enrichment of genes associated with differentially CG methylated regions (0.5 ng sample).
Supplementary Table 10
List of differentially expressed genes in fractions of nuclei extracted from mouse cerebellum.
Supplementary Table 12
List of genes associated with differentially CG-methylated regions in samples from the frontal cortex of clozapine-treated and control mice.
Rights and permissions
About this article
Cite this article
Ma, S., de la Fuente Revenga, M., Sun, Z. et al. Cell-type-specific brain methylomes profiled via ultralow-input microfluidics. Nat Biomed Eng 2, 183–194 (2018). https://doi.org/10.1038/s41551-018-0204-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-018-0204-3
This article is cited by
-
Advances in single-cell omics and multiomics for high-resolution molecular profiling
Experimental & Molecular Medicine (2024)
-
Droplet-based bisulfite sequencing for high-throughput profiling of single-cell DNA methylomes
Nature Communications (2023)
-
Temporally divergent regulatory mechanisms govern neuronal diversification and maturation in the mouse and marmoset neocortex
Nature Neuroscience (2022)
-
Single-cell analysis targeting the proteome
Nature Reviews Chemistry (2020)
-
Microfluidic techniques for enhancing biofuel and biorefinery industry based on microalgae
Biotechnology for Biofuels (2019)