Abstract
Tracking progress towards Target 6.1 of the United Nations Sustainable Development Goals, “achieving universal and equitable access to safe and affordable drinking water for all”, necessitates the development of simple, inexpensive tools to monitor water quality. The rapidly growing field of synthetic biology has the potential to address this need by isolating DNA-encoded sensing elements from nature and reassembling them to create field-deployable “biosensors” that can detect pathogenic or chemical water contaminants. Here, we describe current water quality monitoring strategies enabled by synthetic biology and compare them to previous approaches used to detect three priority water contaminants (i.e., fecal pathogens, arsenic, and fluoride), as well as explain the potential for engineered biosensors to simplify and decentralize water quality monitoring. We conclude with an outlook on the future of biosensor development, in which we discuss their adaptability to emerging contaminants (e.g., metals, agricultural products, and pharmaceuticals), outline current limitations, and propose steps to overcome the field’s outstanding challenges to facilitate global water quality monitoring.
Similar content being viewed by others
Introduction
Reliable access to clean drinking water is essential for human well-being, economic development, and political stability. Impaired water quality, quantity, and accessibility, however, are projected to increase both in frequency and severity due to population increase, climate change, persistent water infrastructure degradation, and poor water governance1,2,3,4,5. As such, institutions like the World Economic Forum6 and the US Government7 have identified the burgeoning water crisis as a top global threat that may undermine progress in protecting human health and serve as a structural driver of poverty and inequity.
The turn of the millennium saw the creation of the United Nations (UN) Millennium Development Goals—eight humanitarian grand challenges to be resolved by 20158. These goals were monitored and refined over the next 15 years9, and after an extensive revision process, 2016 saw the launch of the Sustainable Development Goals (SDGs) for 2030, each of which is accompanied by targets and progress indicators. Sustainable Development Goal (SDG) 6 aspires to “the availability and sustainable management of water and sanitation for all,” with SDG Target 6.1 seeking to “achieve universal and equitable access to safe and affordable drinking water for all”. Progress towards SDG 6.1 is tracked by Indicator 6.1.1, “the proportion of population using safely managed drinking water services,” defined as services that are located on premises, available when needed, and free from contamination10. The Joint Monitoring Program (JMP), housed within the United Nations Children’s Fund (UNICEF) and the World Health Organization (WHO), is the official UN mechanism that has been tasked with monitoring progress towards this goal11.
Accurate tracking and surveillance of global drinking water sources will require significant advances in water quality monitoring technology12,13. Although location on premises and availability when needed can be relatively easily quantified, objectively determining drinking water safety (i.e., if a source is “safely managed”) necessitates the use of technologies to detect the presence of specific contaminants. There are countless potential contaminants that could pose health risks; JMP focuses on three that are globally prevalent and universally recognized as deleterious to human health: arsenic and fluoride (naturally abundant chemical contaminants), and Escherichia coli (an indicator of fecal contamination)14.
Owing to the ubiquity of these contaminants and resource limitations in most affected areas, ideal technologies for global water quality monitoring should be inexpensive, simple enough for an untrained individual to use, and capable of rapidly (within minutes to hours) providing results onsite. Notably, they do not necessarily need to be quantitative; the ability to determine if a contaminant is above or below a risk threshold can provide sufficient actionable information, though technologies that can provide quantitation would enhance their use and impact. Current gold-standard methods for assessing water quality, however, do not fulfill these criteria. Most technologies require expensive equipment and reagents, reliable electricity sources, technically skilled operators, and transportation infrastructure15. For example, the equipment to run qPCR (a DNA amplification technique for pathogen detection) and mass spectrometry (a molecular analysis technique for chemical detection) costs tens of thousands of dollars excluding operational expenses, must be operated by a trained technician, and cannot be brought into the field, thus necessitating sample transport for centralized analysis. As such, these methods come at a significant resource burden, which prohibits widespread deployment16.
While there has been progress in developing more user-friendly field kits capable of rapidly detecting even trace contaminant levels, there is still significant work to be done before they can be widely adopted for global monitoring or individual use3,12. Existing field kits frequently require sample processing steps that are beyond the skill level of an untrained user, along with expensive supplemental equipment or consumables, which are often hazardous chemicals17,18,19,20. Collectively, these limitations preclude the scale and frequency of monitoring that is needed to effectively track progress towards SDG 6.1. There is thus an urgent unmet need for low-cost, field-deployable water quality tests, as evidenced by the UN High Level Panel on Water’s call for higher resolution data on water quality to better address the global water crisis3.
The growing field of synthetic biology, which centers around the design and construction of biological systems21, is poised to address this knowledge gap by engineering and repurposing microbial biosensors. In nature, microbes use biosensors to detect and respond to changes in their environment. For example, a biosensor for detecting toxins may activate the production of proteins that export, neutralize, or metabolize the toxins22. By deconstructing and modifying naturally occurring microbial biosensors, we can create synthetic, genetically encoded biosensors (henceforth referred to as “biosensors”) to detect targets of global concern23. Biosensors have already been developed to detect a wide range of chemicals24,25,26,27,28,29,30,31,32,33,34,35, as well as bacterial36,37,38,39,40,41 and viral42,43,44,45,46,47,48 pathogens. Recently reported biosensors have even been packaged in handheld, easy-to-use formats, facilitating widespread field deployment25,28,40.
Because of their potential to significantly advance the field of water quality monitoring, we seek to provide a primer on emerging biosensors. We specifically focus on the development of field-deployable biosensors42—inexpensive, portable tools that can be used onsite by individuals without technical expertise. While we focus solely on purely genetically encoded biosensors in this review, we note there exist other developing biosensors that are not purely genetically encoded that are covered in other excellent reviews49,50. We begin with a conceptual overview of how the gene expression process can be leveraged for biosensing and discuss the design process for a biosensor. We then discuss their potential applications for detecting Escherichia coli, arsenic, and fluoride, as well as other emerging targets including metals and pharmaceuticals. We conclude with an outlook on the future of synthetic biology for water quality monitoring, identifying needs in the field and necessary steps for widespread implementation.
Biosensor design and construction
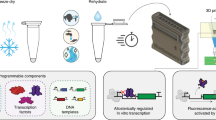
At the core of synthetic biology is the idea that biological systems can be deconstructed into sets of biological parts, which are individual biomolecules with discrete functions (Box 1)23. Each part is written into DNA, which serves as a genetic blueprint. Once they are written into DNA, the individual parts can be manufactured and assembled into a larger functional system (Fig. 1). Synthetic biology works to construct new DNA blueprints that repurpose and reengineer existing biological parts to produce technologies for high-value applications, such as manufacturing food51 and fuels52, creating medicines52, and developing diagnostics53.
A sensor is a biomolecule that recognizes a specific target chemical or fragment of a pathogen’s genome. This recognition event activates cellular machinery (gray), which uses gene expression to generate an output signal in the form of a reporter RNA or protein to indicate the presence of the contaminant. When engineering a biosensor, the sensor and reporter are combined in either an engineered cell or a cell-free system that supports the biological reactions necessary to generate a signal.
Biosensors are molecular systems that detect and respond to specific targets. All biosensors are constructed from two modular parts—a sensor and a reporter54. First, the sensor recognizes a target of interest. Once recognized, the sensor changes its shape to initiate production of the reporter. The reporter then creates a detectable output, typically in the form of fluorescence or color change.
Natural biosensors govern a microbe’s interaction with its environment and exist for every molecule that microbes naturally sense and respond to. They also serve as a starting point for building biosensors, which can be designed, evolved, and engineered to detect targets of interest. To create a biosensor, the DNA encoding the sensor and the reporter is placed in a platform supporting biological function—typically a live cell or cell-free solution containing the cellular machinery needed for transcription and translation.
This review focuses on biosensors for their potential as low-cost, rapid, and field-deployable water quality monitoring devices. Here, we discuss each component of a biosensor and the overall design process for building them.
Sensor parts
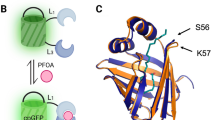
Sensor parts are molecules that detect a target compound54. These molecules can be either natural or engineered and are most commonly nucleic acids or proteins that fold into intricate shapes to match the physical and chemical properties of their targets. One challenge with harvesting parts from nature, however, is that their sensitivity (i.e., ability to detect a particular concentration) or specificity (i.e., ability to distinguish a target from other surrounding molecules) may not meet the requirements for a given application. For example, some natural metal sensors interact with multiple metal ions rather than a single-specific target55. Fortunately, these properties can be adjusted through biomolecular engineering approaches that change the underlying molecular shape and chemistry of the sensor to match the desired sensitivity or specificity56,57.
Reporter parts
Reporter parts are molecules that produce detectable signals. Like sensor parts, they can be nucleic acids or proteins, but they are more varied in their modes of operation because of their range of potential outputs. Fluorescent reporters, the most commonly used reporter type, are molecules that produce a fluorescent signal when illuminated by specific wavelengths of light58. They come in a range of colors and generally require external illumination to visualize their fluorescence. In contrast, colorimetric reporters are enzymes that react with a supplied substrate to produce a visible color. While colorimetric reporters do not require additional equipment to interpret their results, their outputs are more difficult to quantify than fluorescent outputs. Although other reporters that produce luminescent59 or electrical49 outputs exist, we focus on fluorescent and colorimetric reporters given their widespread use in the majority of reported biosensors, as well as their relative field-friendliness.
Biosensor platforms
Biosensors must be housed in a platform that supports the biological processes needed for them to operate. For many sensors, this requires an environment that can support transcription and translation. There are two main biosensing platforms enabling this: “whole-cell” biosensors, which are live cells genetically modified to express the sensor and reporter, and “cell-free” biosensors, which consist of either cellular extract or purified cellular machinery that can perform the processes of gene expression. While both of these platforms are amenable to field deployment because they can be freeze-dried for transport and long-term storage25,28,40,43,44, they each have individual strengths and weaknesses that must be considered during biosensor design.
Whole-cell biosensors have several important advantages. As living sensors, they can be simply and inexpensively mass-produced by allowing the engineered microbe to multiply. They also better replicate the cellular environment that the sensors evolved to function in.
Use of live hosts, however, also presents several challenges60. For instance, whole-cell biosensors must be kept alive during use, requiring bacterial growth media and potentially a field-deployable incubator, which increases the amount of supplemental equipment that must be brought into the field. Furthermore, whole-cell biosensors can only detect targets that do not kill the cell. The synthetic DNA engineered into the cell may also mutate or be lost as cells grow and divide, preventing or distorting sensor and reporter production. Furthermore, the use of live cells inherently confers biocontainment concerns, though methods to encapsulate61 or disable62 whole-cell sensors are being explored to mitigate this risk.
Cell-free biosensors aim to emulate the cellular environment in a non-living system. Placing biosensor DNA in a cell-free gene expression reaction allows the system to act in much the same way as a whole-cell biosensor, but without the complications of needing to maintain and contain living cells. Cell-free biosensors can also be easily tuned and optimized by changing the concentration of the biosensor DNA or other reaction components, which is more difficult to do in a living cell63. Additionally, because some of the physical and biological constraints of live cells are removed, such as the cell’s outer membrane that restricts the import of some targets, they can detect a broader range of contaminants.
These advantages are counterbalanced by the fact that it is difficult to use a part’s performance in a live cell to predict its function in a cell-free platform. Indeed, many sensors require assistance from pieces of peripheral cellular machinery to function properly and the exact differences between the composition of a cell-free reaction and a live cell are still unclear. Despite this, significant progress has been made towards optimizing cell-free systems to accept sensor parts63.
Interaction between sensors and reporters
When placed in a biosensing platform, a sensor controls the activation of a reporter by suppressing its signal until the sensor recognizes its target. As the reporter is only produced when this recognition occurs, the reporter’s signal indicates that the sensor’s target is present. This interaction between a paired sensor and reporter is guided by the way that they are written into the biosensor’s DNA blueprint. For example, protein-based sensors can bind to specific regions of DNA to physically block production of the reporters they regulate, attaching or releasing based on the presence of their target25. In contrast, RNA-based sensors can fold into different shapes based on whether or not a target is present, with different configurations allowing or preventing reporter production28. This diversity of sensor and reporter functions, combined with the staggering number of possible sensor-reporter pairs, offers a vast design space to detect nearly any water contaminant of interest.
Pathogen detection
Waterborne pathogens, including bacteria, protozoa, and viruses, are leading causes of poor water quality globally64 that pose both immediate and long-term risks to human health65. As such, they are currently amongst the highest priority contaminants of global concern16. Fortunately, every pathogenic organism has a unique genetic sequence, which serves as a DNA “barcode” that can be used to identify a specific species and strain in a biosensing reaction. The first step of pathogen detection is sample preparation, where pathogens are broken open to expose their DNA barcodes. These unique DNA sequences are then processed in two steps: amplification of a targeted DNA sequence and production of a signal in response to its detection. This is quite different from existing field-deployable methods that detect secondary indicators of pathogen presence such as H2S production from bacterial metabolism66,67, presence of indicator protein activity19,68,69,70, or biomolecule fluorescence18,71. While these methods are powerful tools for pathogen detection that are currently in use, target DNA sequence detection enables specific pathogen identification, which can provide additional information on water quality and guide treatment more accurately.
There are three basic steps for detecting waterborne pathogens with a biosensor (Fig. 2). While these steps are discussed in the context of detecting fecal coliforms and compared to existing field-deployable coliform detection methods (Table 1), they can be reconfigured to detect virtually any pathogen.
Target amplification
Pathogenic DNA in contaminated water is typically only present in trace amounts. To maximize sensitivity, pathogen detection techniques require some form of amplification to increase the amount of target DNA in a sample. Most synthetic biology approaches use isothermal amplification strategies72, where DNA is amplified while being held at a single temperature. These methods use some of the natural biological machinery used for DNA and RNA replication; by targeting this machinery towards specific sequences in the genome, it is possible to selectively amplify them for detection. These techniques can therefore be made to be highly specific by targeting the unique barcode regions of specific pathogens.
Each isothermal amplification method differs in temperature and time, although most can bring their targets to detectable levels within two hours72. These methods also require minimal training and infrastructure: a freeze-dried reaction containing the biological parts needed for isothermal amplification can be taken to the sample site then deployed by rehydrating with a minimally processed (e.g., syringe-filtered) water sample and incubating, in some cases with body heat or at room temperature. This simplicity eliminates the need for expensive equipment and training, easing deployment in remote and resource limited areas. Collectively, these methods enable detection of even the most dilute pathogens, with methods reporting up to attomolar sensitivity—<10 molecules of DNA in a 10 µL test sample41.
Signal production
The simplest method for pathogenic DNA detection uses modified DNA molecules that produce a fluorescent output in the presence of the target sequence. As DNA is double-stranded, two interacting strands can be attached to a single modified DNA molecule; a fluorescent molecule called a fluorophore is attached to the first strand, while a quencher that inhibits its fluorescent signal is attached to the second. As isothermal amplification creates more target DNA, the amplified DNA displaces the quenching strand to generate a fluorescent output73 (Fig. 2). This method has recently been used to detect as few as 10 contaminating E. coli cells in a 50 mL water sample, with a total assay time of 80 min40. Recent approaches have further built on this strategy to design RNA-based biosensors that undergo similar structural changes, but activate the expression of a reporter gene in the presence of specific bacterial or viral DNA barcodes37,43.
Beyond its widely known uses for gene editing, Clustered Regularly Interspaced Short Palindromic Repeats, or CRISPR, provides a powerful new method for pathogen detection. CRISPR systems are hybrid protein-RNA biosensors. In CRISPR, portions of a special “guide” RNA target DNA barcode sequences of interest, which are then destroyed by an associated CRISPR protein. In a biosensor, CRISPR systems can be used alongside DNA or RNA sequences labeled by a fluorophore-quencher pair to produce a detectable signal upon target recognition (Fig. 2)46,74. Using this strategy, recently developed CRISPR-based sensors have reached the maximum possible specificity by discriminating between pathogenic DNA sequences that differ by only a single base pair75.
Outstanding challenges for pathogen detection
Biosensors target barcode sequences in genomic DNA, which is protected by the cell’s outer wall and therefore inaccessible in an unprepared sample. Because of this, they require some means to break open cells and access their DNA. This can be difficult to do in the field, although some technologies are beginning to address this limitation46. Another limitation is that these technologies cannot distinguish between live and dead pathogens; DNA from dead pathogens remains detectable in water for days before it degrades76, which means that a positive result is not a perfect indicator of water quality. Lastly, while the use of isothermal amplification allows for detection of trace amounts of pathogen DNA, the process of amplification obscures the pathogen’s original concentration and hampers precise quantification. Several existing strategies use mathematical models to infer pathogen concentration from final fluorescent signal strength37,75, but precise quantification will likely require sophisticated peripheral equipment or an array of tests with built-in thresholds.
Chemical contaminant detection
Some of the most significant threats to our water supply are chemical contaminants—molecules that are deleterious to human health when consumed at dangerous levels. Many of those compounds occur naturally in soil, while others enter the water supply from industrial pollution, agricultural runoff, or deficient utility infrastructure. Sensors for chemical contaminants do not require an amplification step and therefore work more similarly to natural biosensors than pathogen biosensors do. There are two steps to detecting a chemical contaminant: the biosensor first recognizes its target chemical, which in turn initiates production of a reporter that generates a detectable signal. Currently, significant progress has been made in developing biosensors to detect arsenic33 and fluoride28, two of the WHO’s highest-priority chemical water contaminants16. Here, we discuss recent progress towards using biosensors for chemical sensing with a focus on arsenic and fluoride, comparing these tools to existing field-deployable methods (Table 1).
Arsenic
Arsenic contamination of groundwater is typically caused by the leaching of naturally occurring arsenous compounds from the surrounding soil77. Consumption of arsenic-contaminated water is associated with lesions, cardiovascular and pulmonary disease, and cancer in humans78. Current field-deployable methods for arsenic detection utilize a colorimetric chemical test strip to semi-quantitatively detect as low as 5 ppb arsenic within a few minutes20. However, these tests require significant technical skill from the user and produce toxic byproducts, such as arsine gas.
Published biosensors for arsenic are generally whole-cell sensors that are controlled by an arsenic-responsive protein79 (Fig. 3a). In the absence of arsenic, the protein binds to the biosensor DNA, stopping the reporter from being produced. Arsenic binds to the protein and causes it to change shape, releasing the biosensor DNA and allowing production of the reporter. Previous arsenic biosensors have used fluorescent, colorimetric, and luminescent outputs, with reported detection as low as single-digit parts per billion79. This offers presence/absence results for arsenic concentrations below the 10 ppb WHO guideline for arsenic in drinking water16, though these sensors have yet to be extensively validated in field conditions79.
a Detection of arsenic using a protein sensor in a whole-cell biosensor. Once the protein sensor recognizes the ligand arsenic, it releases reporter DNA and allows a reporter molecule such as a fluorescent protein to be produced. b Detection of fluoride using an RNA sensor in a cell-free biosensor. The RNA sensor recognizes the ligand fluoride and changes its shape to allow the production of a reporter molecule. The specific reporter molecule shown is an enzyme that can convert a colorless substrate into a yellow substance.
Fluoride
Fluoride leaches into groundwater from naturally occurring soil minerals and can also be introduced by agricultural runoff or the precipitation of fluoride-containing industrial ash in rainwater80. Chronic consumption of fluoride-contaminated water causes dental and skeletal fluorosis, which manifests as discolored teeth, weakened bones, seizures, and stunted growth80. Current field-deployable methods for fluoride detection utilize either photometric analytical equipment to semi-quantitively measure a colorimetric reaction17 or a quantitative fluoride-sensing electrode81. While both of these methods can safely detect down to 0.1 ppm fluoride, they require expensive supplementary equipment for their use, precluding widespread deployment.
A recently developed biosensor for fluoride uses a naturally occurring RNA regulator, called a riboswitch, in a cell-free system28 (Fig. 3b). In the absence of fluoride, the riboswitch folds into a structure that stops the reporter from being produced. When present, fluoride ions bind to the riboswitch, causing it to fold into an alternate structure that permits production of the reporter. This fluoride biosensor can be paired to both fluorescent and colorimetric outputs, with reported detection as low as one part per million28. During preliminary field testing28, this sensor provided presence/absence results in environmental samples below the 1.5 ppm WHO guideline for fluoride in drinking water16.
Emerging contaminants
Biosensors also have the potential to detect emerging contaminants beyond arsenic and fluoride, including metals, agricultural products, and pharmaceutical and personal care products (PPCPs), such as antibiotics and cosmetics. Both whole-cell and cell-free biosensors have previously been used to detect metals by utilizing natural or engineered proteins; sensors have been reported for cadmium, lead, mercury, arsenic, copper, zinc, nickel, and cobalt, with sensitivities ranging from low parts per million to parts per billion25,31,82,83. Sensors for atrazine, a toxic herbicide, have also been developed by encoding a natural metabolic pathway for atrazine’s conversion to cyanuric acid, which can be detected with a known protein sensor84,85. Furthermore, new cell-free approaches can detect a range of PPCPs, including multiple families of antibiotics and benzalkonium chloride25,82,86. The ability to detect such a wide range of targets underscores the potential of biosensors as modular chemical sensing platforms, paving the way for rapid sensor development and deployment to detect new and emerging contaminants of concern.
Outstanding challenges for chemical contaminant detection
While many whole-cell sensors report WHO-relevant limits of detection, they are limited by the deployment and operational concerns that are characteristic of live cells. Additionally, many of these sensors are susceptible to false positives due to interference by other chemical contaminants and unintended reporter production. To overcome these challenges, substantial progress must be made in developing robust biocontainment strategies and methods to tune biosensor sensitivity and specificity. While cell-free biosensors partially resolve some of these problems by virtue of being non-living, easily tunable systems, it is still difficult to completely predict how tuning certain parameters influences a sensor’s function. Further development of cell-free sensors must therefore focus on identifying the factors that contribute to maximal sensor and reporter function in cell-free systems and optimizing them for biosensing.
Discussion
Infrequent monitoring of a narrow range of contaminants has created significant gaps in our current understanding of water quality12 and therefore water insecurity87,88,89. Synthetic biology has the potential to fill these knowledge gaps by offering simple, field-deployable tools to report on individual water supplies or serve as pre-screening tools to be used with existing gold-standard methods to provide the large-scale, high-resolution data needed to track progress towards development goals. While there are existing field-deployable tools, they are limited by the need for the technical expertise, supplemental equipment, or dangerous chemical reagents required for their use (Table 1). The potential for biosensors to decrease cost and improve ease-of-use for such diagnostics relative to current methods would enable more frequent measurements across wider and more diverse regions, producing water quality data that are more comprehensive and specific than currently available. There is significant promise for this to become a reality—current biosensor formats are accessible to an untrained user, and recent cost estimates suggest that their production can be scaled for global use. Freeze-dried, cell-free reactions can currently be manufactured for a few cents per sensor, with even lower costs possible for whole-cell biosensors90. Moving forward, these costs could decrease by as much as one order of magnitude90, further facilitating mass deployment.
This potential is counterbalanced by several existing barriers to rapid biosensor design and deployment. For example, we are currently limited to harvesting sensor parts from nature, rather than designing them from scratch. With the rise of unnatural contaminants such as synthetic antibiotics, pesticides, and other harmful industrial compounds, we may lack the tools to detect some emerging targets. While we are currently on the cusp of engineering entirely synthetic proteins91 and RNAs92 to address this need, the technologies to do so are still in their infancy, and it will be some time before they can be applied to targeted contaminant detection.
There is also significant work to be done in developing validated field deployment strategies. Although many biosensors can be freeze-dried for transport and long-term storage25,28,40,43,44, this has not yet been explored in the context of tools to enable their use in real-world settings. Of particular concern is the potential for other compounds present in environmental samples to interfere with biosensor components, or for organic matter to chelate contaminants and mask their presence. Thus, a major next step for biosensor development is to characterize these potential inhibitory effects and devise strategies to make biosensors robust against them83.
Fortunately, there have already been several successes using these biosensors in complex samples. For example, cell-free biosensors have been used in the laboratory to detect fecal contamination of unprocessed water samples, including a test for robustness with raw sewage40. Additionally, a cell-free fluoride biosensor was capable of detecting environmental fluoride in unprocessed water samples onsite28. While these preliminary findings suggest that cell-free biosensors can be robust to a wide range of potential contaminants in complex water samples, more exhaustive testing that considers the effect that common contaminants and other compounds found at target sites is needed. We must also develop comprehensive packaging and usage guidelines that accommodate both the needs and abilities of diverse users.
From a logistical perspective, the lengthy validation and regulatory approval processes for certifying biosensors may delay their potential impact. Furthermore, meeting the manufacturing demands for global deployment will require funding and production capabilities beyond the reach of academic labs. Enabling individuals to more easily monitor their own water quality could also reveal unwelcome information, and may raise some potential societal, ethical, data protection, and regulatory concerns. These issues will require the careful consideration and cooperation of diverse stakeholders to ensure that these technologies are used for the maximal public good. Because of this, widespread implementation of these technologies will require interdisciplinary collaboration across the synthetic biology and water, sanitation, and hygiene (WASH) communities to foster the use of biological design to advance large-scale humanitarian goals.
As our ability to build biological systems improves, we can begin constructing more sophisticated systems from a wider array of biological parts. Recent work has demonstrated that biosensors can do more than merely produce a single reporter output in response to a target. Indeed, networks of interacting genes can be coupled to form “molecular computers” that take input signals from a biosensor and calculate an appropriate response93. For example, a genetic system could be engineered to simultaneously detect multiple targets and produce an output that reports the identity and concentration of each target. Furthermore, new sample calibration strategies83 are being developed to circumvent biosensors’ intrinsic limitations and enable field-deployable sample quantification. As we continue harvesting parts from nature and clarifying biological design principles, we expect to see an improvement in the sensitivity and specificity of biosensors for an expanding list of detectable targets.
Using biosensors to generate spatiotemporal water quality data will enable more efficient resource allocation by showing exactly when and where interventions are necessary. Not only will such diagnostics provide important population-level information, but they have the potential to usher in the ability to simply and inexpensively assess water quality so that even untrained individuals can personally test the safety of their water. As such, advances in synthetic biology could facilitate global water quality monitoring by producing actionable contaminant data, guiding the development of efficacious policies and programs, and informing choices about the water we consume.
References
Gosling, S. N. & Arnell, N. W. A global assessment of the impact of climate change on water scarcity. Clim. Change 134, 371–385 (2016).
Kummu, M. et al. The world’s road to water scarcity: shortage and stress in the 20th century and pathways towards sustainability. Sci. Rep. 6, 38495 (2016).
United Nations. High-Level Panel on Water. Making Every Drop Count: An Agenda for Water Action. https://reliefweb.int/report/world/making-every-drop-count-agenda-water-action-high-level-panel-water-outcome-document-14 (2018).
Watkins, K. Human Development Report 2006-beyond Scarcity: Power, Poverty and the Global Water Crisis. (UNDP Human Development Reports, 2006).
Honkonen, T. Water security and climate change: the need for adaptive governance. Potchefstroom Electron. Law Journal/Potchefstroomse Elektron. Regsbl. 20. https://doi.org/10.17159/1727-3781/2016/v19i0a1651 (2017).
World Economic Forum. The Global Risks Report 2019, 14th Edition. https://www.weforum.org/reports/the-global-risks-report-2019 (2019).
USAID. U.S. Government Global Water Strategy. https://www.usaid.gov/what-we-do/water-and-sanitation/us-global-water-strategy (USAID, 2017).
Wagstaff, A. The Millennium Development Goals for Health: Rising to the Challenges. http://documents.worldbank.org/curated/en/875031468329973611/The-millennium-development-goals-for-health-rising-to-the-challenges (2004).
Sachs, J. D. From millennium development goals to sustainable development goals. Lancet 379, 2206–2211 (2012).
World Health Organization & UNICEF. Water for Life: Making It Happen. https://www.who.int/water_sanitation_health/monitoring/jmp2005/en/ (2005).
Bartram, J. et al. Global monitoring of water supply and sanitation: history, methods and future challenges. Int. J. Environ. Res. Public Health 11, 8137–8165 (2014).
Damania, R., Desbureaux, S., Rodella, A.-S., Russ, J. & Zaveri, E. Quality Unknown: The Invisible Water Crisis. (World Bank Publications, 2019).
Stauber, C. The sustainable development goals for water: the need to consider perception, preference, and safety. Am. J. Trop. Med. Hyg. 97, 985 (2017).
UNICEF. SDG Global Indicators Related to Children. https://data.unicef.org/resources/sdg-global-indicators-related-to-children/ (2018).
Ellis, K. et al. Understanding the costs of investigating coliform and E. coli detections during routine drinking water quality monitoring. Urban Water J. 15, 101–108 (2018).
World Health Organization. Guidelines for Drinking-water Quality, 4th edn, Incorporating the 1st Addendum. (World Health Organization, 2018).
Fawell, J., Bailey, K., Chilton, J., Dahi, E. & Magara, Y. Fluoride in Drinking-Water. (IWA Publishing, 2006).
Chelsea Technologies. UviLux. Available at: https://chelsea.co.uk/products/uvilux/.
IDEXX. Coliform/E.coli Water Tests. Available at: https://www.idexx.com/en/water/products/?cy=y_category_252&cx=x_category_259&ts=all.
Hach USA. Arsenic Low Range Test Kit. Available at: https://www.hach.com/arsenic-low-range-test-kit/product?id=7640217303.
Khalil, A. S. & Collins, J. J. Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367 (2010).
Liu, X. et al. Selective cadmium regulation mediated by a cooperative binding mechanism in CadR. Proc. Natl Acad. Sci. 116, 20398–20403 (2019).
Lucks, J. B., Qi, L., Whitaker, W. R. & Arkin, A. P. Toward scalable parts families for predictable design of biological circuits. Curr. Opin. Microbiol. 11, 567–573 (2008).
Salehi, A. S. M. et al. Cell-free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal. Chem. 89, 3395–3401 (2017).
Alam, K. K. et al. Rapid, low-cost detection of water contaminants using regulated in vitro transcription. Preprint at biorxiv.org/content/10.1101/619296v2 (2019).
Willardson, B. M. et al. Development and testing of a bacterial biosensor for toluene-based environmental contaminants. Appl. Environ. Microbiol. 64, 1006–1012 (1998).
Tauriainen, S., Karp, M., Chang, W. & Virta, M. Luminescent bacterial sensor for cadmium and lead. Biosens. Bioelectron. 13, 931–938 (1998).
Thavarajah, W. et al. Point-of-use detection of environmental fluoride via a cell-free riboswitch-based biosensor. ACS Synth. Biol. 9, 10–18 (2019).
Selifonova, O., Burlage, R. & Barkay, T. Bioluminescent sensors for detection of bioavailable Hg (II) in the environment. Appl. Environ. Microbiol. 59, 3083–3090 (1993).
Gräwe, A. et al. A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. PLoS ONE 14, e0210940 (2019).
Pellinen, T., Huovinen, T. & Karp, M. A cell-free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter. Anal. Biochem. 330, 52–57 (2004).
Rawson, D. M., Willmer, A. J. & Turner, A. P. P. Whole-cell biosensors for environmental monitoring. Biosensors 4, 299–311 (1989).
Stocker, J. et al. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 37, 4743–4750 (2003).
Prest, A. G., Winson, M. K., Hammond, J. R. M. & Stewart, G. The construction and application of a lux‐based nitrate biosensor. Lett. Appl. Microbiol. 24, 355–360 (1997).
Virta, M., Lampinen, J. & Karp, M. A luminescence-based mercury biosensor. Anal. Chem. 67, 667–669 (1995).
Wen, K. Y. et al. A cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa-infected respiratory samples. ACS Synth. Biol. 6, 2293–2301 (2017).
Takahashi, M. K. et al. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 9, 3347 (2018).
Phillips, E. A., Moehling, T. J., Bhadra, S., Ellington, A. D. & Linnes, J. C. Strand displacement probes combined with isothermal nucleic acid amplification for instrument-free detection from complex samples. Anal. Chem. 90, 6580–6586 (2018).
Kawaguchi, T., Chen, Y. P., Norman, R. S. & Decho, A. W. Rapid screening of quorum-sensing signal N-acyl homoserine lactones by an in vitro cell-free assay. Appl. Environ. Microbiol. 74, 3667–3671 (2008).
Jiang, Y. S. et al. Portable platform for rapid in-field identification of human fecal pollution in water. Water Res. 131, 186–195 (2018).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science (80-.). 356, 438–442 (2017).
Pardee, K. et al. Paper-based synthetic gene networks. Cell 159, 940–954 (2014).
Pardee, K. et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016).
Verosloff, M., Chappell, J., Perry, K. L., Thompson, J. R. & Lucks, J. B. PLANT-Dx: a molecular diagnostic for point-of-use detection of plant pathogens. ACS Synth. Biol. 8, 902–905 (2019).
Ma, D., Shen, L., Wu, K., Diehnelt, C. W. & Green, A. A. Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment. Synth. Biol. 3, ysy018 (2018).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science (80-.). 360, 444–448 (2018).
Spoelstra, W. K. et al. CRISPR-based DNA and RNA detection with liquid-liquid phase separation. Preprint at biorxiv.org/content/10.1101/471482v2 (2019).
Romero, J. L. R., Carver, G. D., Johnson, P. A., Perry, K. L. & Thompson, J. R. A rapid, sensitive and inexpensive method for detection of grapevine red blotch virus without tissue extraction using loop-mediated isothermal amplification. Arch. Virol. 164, 1453–1457 (2019).
Grieshaber, D., MacKenzie, R., Vörös, J. & Reimhult, E. Electrochemical biosensors-sensor principles and architectures. Sensors 8, 1400–1458 (2008).
Wang, J. et al. DNA electrochemical biosensors for environmental monitoring. A review. Anal. Chim. Acta 347, 1–8 (1997).
Tyagi, A. et al. Synthetic biology: applications in the food sector. Crit. Rev. Food Sci. Nutr. 56, 1777–1789 (2016).
Mortimer, J. C. Plant synthetic biology could drive a revolution in biofuels and medicine. Exp. Biol. Med. 244, 323–331 (2019).
Slomovic, S., Pardee, K. & Collins, J. J. Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl Acad. Sci. 112, 14429–14435 (2015).
Rogers, J. K. et al. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 43, 7648–7660 (2015).
Porto, T. V., Wilson, M. T. & Worrall, J. A. R. Copper and nickel bind via two distinct kinetic mechanisms to a CsoR metalloregulator. Dalt. Trans. 44, 20176–20185 (2015).
Yuan, L., Kurek, I., English, J. & Keenan, R. Laboratory-directed protein evolution. Microbiol. Mol. Biol. Rev. 69, 373–392 (2005).
Porter, E. B., Polaski, J. T., Morck, M. M. & Batey, R. T. Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nat. Chem. Biol. 13, 295 (2017).
Naylor, L. H. Reporter gene technology: the future looks bright. Biochem. Pharmacol. 58, 749–757 (1999).
Thorne, N., Inglese, J. & Auld, D. S. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol. 17, 646–657 (2010).
Wan, X., Ho, T. Y. H. & Wang, B. in Handbook of Cell Biosensors. 1–37 (2019).
Belkin, S. et al. Remote detection of buried landmines using a bacterial sensor. Nat. Biotechnol. 35, 308 (2017).
Stirling, F. et al. Rational design of evolutionarily stable microbial kill switches. Mol. Cell 68, 686–697 (2017).
Silverman, A., Kelley-Loughnane, N., Lucks, J. B. & Jewett, M. C. Deconstructing cell-free extract preparation for in vitro activation of transcriptional genetic circuitry. ACS Synth. Biol. 8, 403–414 (2018).
Pandey, P. K., Kass, P. H., Soupir, M. L., Biswas, S. & Singh, V. P. Contamination of water resources by pathogenic bacteria. AMB Express 4, 51 (2014).
Jamison, D. T. et al. Disease Control Priorities, (Volume 9): Improving Health and Reducing Poverty. (World Bank Publications, 2017).
Pillai, J., Mathew, K., Gibbs, R. & Ho, G. E. H2S paper strip method—A bacteriological test for faecal coliforms in drinking water at various temperatures. Water Sci. Technol. 40, 85–90 (1999).
Aquagenx. The Compartment Bag Test (CBT) Quantitative H2S Water Quality Test Kits. Available at: https://www.aquagenx.com/cbt-h2s/.
Cowburn, J. K., Goodall, T., Fricker, E. J., Walter, K. S. & Fricker, C. R. A preliminary study of the use of Colilert for water quality monitoring. Lett. Appl. Microbiol. 19, 50–52 (1994).
mWater. mWater Test Kits. Available at: https://www.mwater.co/kits.html.
Pall Corporation. Membrane Filter Technique. Available at: https://laboratory.pall.com/en/microbiology-qc/laboratory-membrane-filter-technique.html.
Sorensen, J. P. R. et al. In-situ tryptophan-like fluorescence: a real-time indicator of faecal contamination in drinking water supplies. Water Res. 81, 38–46 (2015).
Zhao, Y., Chen, F., Li, Q., Wang, L. & Fan, C. Isothermal amplification of nucleic acids. Chem. Rev. 115, 12491–12545 (2015).
Kim, Y., Sohn, D. & Tan, W. Molecular beacons in biomedical detection and clinical diagnosis. Int. J. Clin. Exp. Pathol. 1, 105 (2008).
Chen, J. S. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science (80-.). 360, 436–439 (2018).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science (80-.). 360, 439–444 (2018).
Eichmiller, J. J., Best, S. E. & Sorensen, P. W. Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ. Sci. Technol. 50, 1859–1867 (2016).
Nickson, R. et al. Arsenic poisoning of Bangladesh groundwater. Nature 395, 338 (1998).
Hughes, M. F., Beck, B. D., Chen, Y., Lewis, A. S. & Thomas, D. J. Arsenic exposure and toxicology: a historical perspective. Toxicol. Sci. 123, 305–332 (2011).
Kaur, H., Kumar, R., Babu, J. N. & Mittal, S. Advances in arsenic biosensor development–a comprehensive review. Biosens. Bioelectron. 63, 533–545 (2015).
Brindha, K. & Elango, L. Fluoride in groundwater: causes, implications and mitigation measures. Fluoride Prop. Appl. Environ. Manag. 1, 111–136 (2011).
Hanna Instruments. Fluoride Portable Meter. Available at: https://www.hannainst.com/fluoride-portable-meter-hi98402.html.
Didovyk, A., Tonooka, T., Tsimring, L. & Hasty, J. Rapid and scalable preparation of bacterial lysates for cell-free gene expression. ACS Synth. Biol. 6, 2198–2208 (2017).
McNerney, M. P. et al. Point-of-care biomarker quantification enabled by sample-specific calibration. Sci. Adv. 5, eaax4473 (2019).
Hua, A., Gueuné, H., Cregut, M., Thouand, G. & Durand, M. -J. Development of a bacterial bioassay for atrazine and cyanuric acid detection. Front. Microbiol. 6, 211 (2015).
Silverman, A. D., Akova, U., Alam, K. K., Jewett, M. C. & Lucks, J. B. Design and optimization of a cell-free atrazine biosensor. ACS Synth. Biol. https://doi.org/10.1021/acssynbio.9b00388 (2020).
Karig, D. K., Iyer, S., Simpson, M. L. & Doktycz, M. J. Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Res. 40, 3763–3774 (2011).
Jepson, W. E., Wutich, A., Colllins, S. M., Boateng, G. O. & Young, S. L. Progress in household water insecurity metrics: a cross‐disciplinary approach. Wiley Interdiscip. Rev. Water 4, e1214 (2017).
Young, S. L. et al. The Household Water InSecurity Experiences (HWISE) Scale: development and validation of a household water insecurity measure for low-income and middle-income countries. BMJ Glob. Heal. 4, e001750 (2019).
Wutich, A. et al. Advancing methods for research on household water insecurity: studying entitlements and capabilities, socio-cultural dynamics, and political processes, institutions and governance. Water Secur. 2, 1–10 (2017).
Silverman, A. D., Karim, A. S. & Jewett, M. C. Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet. 21, 151–170 (2020).
Huang, P. -S., Boyken, S. E. & Baker, D. The coming of age of de novo protein design. Nature 537, 320 (2016).
Lee, Y. J. & Moon, T. S. Design rules of synthetic non-coding RNAs in bacteria. Methods 143, 58–69 (2018).
Nandagopal, N. & Elowitz, M. B. Synthetic biology: integrated gene circuits. Science (80-.). 333, 1244–1248 (2011).
Alberts, B. et al. Molecular Biology of the Cell 6th edn. (Garland Science, 2015).
Acknowledgements
We would like to acknowledge Adam Silverman (Northwestern University) and Professor Samuel Dorevitch (University of Illinois, Chicago) for helpful comments and discussions in preparing the manuscript. W.T. and M.S.V. were supported in part by the National Institutes of Health Training Grant (T32GM008449) through Northwestern University’s Biotechnology Training Program. This work was also supported by the National Science Foundation (1929912 to J.B.L.), an NSF CAREER award (1452441 to J.B.L.), Searle Funds at the Chicago Community Trust (to J.B.L.), the Andrew Carnegie Foundation (to S.L.Y.), the Air Force Research Laboratory Center of Excellence for Advanced Bioprogrammable Nanomaterials (C-ABN) Grant FA8650-15-2-5518 (to M.C.J. and J.B.L.), the David and Lucile Packard Foundation (to M.C.J.), the Camille Dreyfus Teacher-Scholar Program (to M.C.J. and J.B.L.), and the generous support of the American people provided to the Feed the Future Sustainable Intensification Innovation Lab (SIIL) through the United State Agency for International Development Cooperative Agreement AID-OAA-L-14-00006 (to S.L.Y.). The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the Air Force Research Laboratory, Air Force Office of Scientific Research, the National Science Foundation, USAID, or the United States Government.
Author information
Authors and Affiliations
Contributions
Conceptualization: W.T., K.K.A., S.L.Y., and J.B.L. Project administration: W.T. and J.B.L. Funding acquisition: M.C.J, S.L.Y., & J.B.L. Writing—original draft: W.T., M.S.V., J.K.J., J.D.M., S.L.Y., and J.B.L. Writing—review and editing: W.T., M.S.V., J.K.J., K.K.A., J.D.M., M.C.J., S.L.Y., and J.B.L.
Corresponding authors
Ethics declarations
Competing interests
K.K.A., J.B.L., and M.C.J. have a financial interest in Stemloop, Inc. These latter interests were reviewed and managed by Northwestern University in accordance with their conflict of interest policies. All other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thavarajah, W., Verosloff, M.S., Jung, J.K. et al. A primer on emerging field-deployable synthetic biology tools for global water quality monitoring. npj Clean Water 3, 18 (2020). https://doi.org/10.1038/s41545-020-0064-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41545-020-0064-8
This article is cited by
-
The accuracy and usability of point-of-use fluoride biosensors in rural Kenya
npj Clean Water (2023)
-
Feedforward growth rate control mitigates gene activation burden
Nature Communications (2022)
-
Applications, challenges, and needs for employing synthetic biology beyond the lab
Nature Communications (2021)
-
Drinking water quality and the SDGs
npj Clean Water (2020)