Abstract
Bacterial infection remains as one of the major healthcare issues, despite significant scientific and medical progress in this field. Infection by Streptococcus Pneumoniae (S. Pneumoniae) can cause pneumonia and other serious infectious diseases, such as bacteremia, sinusitis and meningitis. The pneumococcal capsular polysaccharides (CPS) that constitute the outermost layer of the bacterial cell are the main immunogens and protect the pathogen from host defense mechanisms. Over 90 pneumococcal CPS serotypes have been identified, among which more than 30 can cause invasive pneumococcal diseases that could lead to morbidity and mortality. Multivalent pneumococcal vaccines have been developed to prevent diseases caused by S. Pneumoniae. These vaccines employ either purified pneumococcal CPSs or protein conjugates of these CPSs to generate antigen-specific immune responses for patient protection. Serotype-specific quantitation of these polysaccharides (Ps) antigen species are required for vaccine clinical dosage, product release and quality control. Herein, we have developed an antibody-enhanced high-performance liquid chromatography (HPLC) assay for serotype-specific quantitation of the polysaccharide contents in multivalent pneumococcal conjugate vaccines (PCVs). A fluorescence-labeled multiplex assay format has also been developed. This work laid the foundation for a serotype-specific antigen assay format that could play an important role for future vaccine research and development.
Similar content being viewed by others
Introduction
Worldwide, diseases caused by Streptococcus Pneumoniae are responsible for the major proportion of deaths in children under 5 years of age and adults over 50. Patients can be infected by different serotypes of the bacteria simultaneously or at different times, either continuously or intermittently. By far, more than 90 S. Pneumoniae strains have been identified and over 30 of them can cause streptococcus infections1,2. Streptococcus Pneumoniae are encapsulated gram-positive bacteria. Its immunogenic factors largely come from capsular polysaccharides (CPS) that constitute the bacterial cell wall3,4,5,6. Each pneumococcal serotype (ST) infects the host with a specific CPS antigen. Each CPS antigen is a unique polysaccharide (Ps) polymer with its own repeating unit (RU) structure (ST4 example in Supplementary Fig. 1). Conjugation of polysaccharide to a carrier protein, such as CRM197 help to recruit helper T cells, and offer long-lasting and more robust T-cell-dependent immune responses7,8,9,10,11,12. Although two powerful pneumococcal conjugate vaccines (PCVs), PREVNAR 20 and VAXNEUVANC have been approved by FDA recently, there are still great needs for novel pneumococcal vaccines that can offer more robust immune responses and greater serotype coverages13,14,15,16. Analytical methods that monitor critical quality attributes (CQA) in such complex multi-valent vaccine product with increasing serotype repertoire often encounter challenges, but are critical for the success of vaccine development17,18,19.
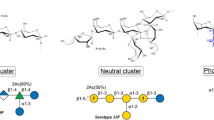
Our 15-valent pneumococcal conjugate vaccine (PCV15) consists of pneumococcal CPS serotypes 1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, 22F, 23F, and 33F. Each CPS is conjugated to the carrier protein CRM197 individually, resulting a monovalent conjugate polysaccharide (conjugated Ps). Residual amount of unconjugated polysaccharide species (free Ps or free polysaccharide) also coexists with conjugated Ps in the drug substance (Fig. 1). Both conjugated Ps and free Ps can elicit serotype-specific immune responses once dosed to the patient, with the conjugate Ps being the more desired antigen form that can elicit long-lasting T-cell-dependent immune responses.
To assess the potency and quality of the vaccine, a serotype-specific polysaccharide quantitation assay will need to be implemented in vaccine development and manufacturing. Conventionally, such serotype-specific antigen assays were performed on the immunoassay platforms; among them, the sandwich enzyme-linked immunosorbent assay (ELISA) is the preferred format for this assay20,21. Although immunoassays, such as ELISA have been reported to quantify antigen content in several vaccines22,23,24, there is only a light scattering (LS)-based nephelometry method reported for pneumococcal vaccines25. Due to the lack of precedent reports for a reliable antigen-specific pneumococcal immunoassay and the need for such assay for our PCV development, we sought to develop an innovative assay platform for serotype-specific polysaccharide quantitation. For the last few decades, high-performance liquid chromatography (HPLC) has been demonstrated as a highly robust and automatic assay platform for vaccine analysis26,27. HPLC methods have also been utilized to report the concentration of a single polysaccharide or total polysaccharides in a sample28,29,30. However, HPLC methods have not been established for serotype-specific analysis for a sample containing multivalent antigens. The polysaccharides in the PCV are large molecular weight (Mw ~100–500 kDa), polydisperse and flexible biopolymers with overlapped monosaccharide components in their repeating unit (RU) structures31,32. Therefore, direct chromatography separation and detection were not able to deconvolute each serotype in a multivalent vaccine.
We proposed that we could establish a novel antibody-enhanced chromatography assay for serotype-specific antigen quantitation by leveraging the specificity of each anti-serotype (anti-ST) antibody (Ab). An anti-ST Ab binds to only its corresponding polysaccharide serotype without cross-reactivity to untargeted serotypes. By complexing of an anti-ST Ab and multi-polysaccharide serotypes together in an antibody–antigen binding reaction, a serotype-specific antibody–polysaccharide complex (APC) would form in solution. The serotype-specific APC can therefore serve as a surrogate to analyze the target polysaccharide serotype. Once the APC is separated from the excess unbound antibody, the APC peak area on the chromatogram (or electropherogram) can be used to quantify the antibody-targeted polysaccharide serotype. The unbound serotypes (untargeted) do not need to be separated from either APC or Ab, since they do not have the chromophore (or fluorophore) used for detection. Only species that hold antibody (either APC or antibody itself) are visible as peaks on the chromatogram (Fig. 2).
According to structural studies33,34,35,36,37, an anti-serotype antibody binds to a defined polysaccharide structural motif (epitope). The polysaccharide epitope is formed by a defined number of repeating units (RUs). One polysaccharide molecule has multiple antibody-binding epitopes, with each epitope consisting of a defined number of RUs (Fig. 3). When the epitopes on the polysaccharide are saturated by the antibody binding, the number of antibodies on the resulting APC will represent the number of RUs on the polysaccharide, which can be converted to molar concentration of RU (mM), or weight concentration of the polysaccharide (µg/mL) based on the known molar mass (Mw or molecular weight) of the specific RU. Guided by this structural information, we established a serotype-specific assay that quantifies the free polysaccharide content in our PCV15 vaccine.
Each antibody-binding epitope is formed by a number (n = 3–9) of polysaccharide repeating unit (RU). Each RU is formed by a few individual monosaccharides represented by different shapes in the bracket. When all epitopes are saturated by antibody binding, the amount of antibody bound on the polysaccharide can reflect the polysaccharide amount.
Conjugated polysaccharide in a PCV bear a carrier protein that could interfere with antibody-assisted polysaccharide quantitation in its original format. Both antibody and CRM197 have signal at Ex/Em 280 nm/352 nm used for fluorescence detection (or UV 280 nm absorption), since they both are proteins. This conundrum can be resolved by labeling the anti-serotype antibody with a fluorescent tag (FLR tag) and switching the detection channel to the wavelength that is unique to the fluorescent tag, e.g., Ex/Em 433 nm/541 nm, at which unlabeled CRM197 proteins on the conjugate do not have a detectable signal. Hence, the FLR tag-labeled antibody chromatography can be used to quantify the protein-conjugated polysaccharide. Furthermore, simultaneous detections at multiple wavelengths can be easily set up on modern HPLC instruments. When different anti-serotype antibodies are labeled with several distinctive FLR tags, the signal from these preselected FLR tags can be detected and resolved on multidetection channels on a HPLC. Therefore, multiple conjugated polysaccharide serotypes in a complex multivalent vaccine can be analyzed in a single HPLC run (Fig. 4), and a multiplex serotype-specific antigen assay platform was developed.
Results
Development of serotype-specific HPLC assay
Since we had previous successful size-exclusion chromatography (SEC) methods for the vaccine, free Ps, and protein-conjugated Ps characterization and quantitation38,39,40, we decided to start our assay development on the same platform. An anti-ST1 mAb was mixed with pneumococcal serotype 1 (ST1) Ps standard and injected on the SEC-UV-MALS-RI system (“Methods”). A higher Mw peak that was separated from the mAb was observed on all three detection channels, whereas the un-complexed Ps could not be detected on UV A280. This substantiated the formation of antibody–polysaccharide complex (APC), which was detected with the chromophore acquired from the mAb at A280 (Fig. 5 and Table 1).
We then moved to probe the assay linearity range of one serotype with fluorescence (FLR) detection, so we could later expand the assay to remaining serotypes. Under binding conditions with excess mAb vs serotype 4 (ST4) polysaccharide (Ps), the ST4 antibody–polysaccharide complexes (APCs) were observed on the SEC chromatogram at all Ps concentration levels (Fig. 6a), and a linear FLR response curve was established from the polysaccharide (Ps) concentration range of 0.01 to 0.15 µg/mL (Supplementary Table 1). As Ps concentration increased above 0.15 µg/mL, the FLR response curve started to flatten, indicating that the multi-epitopes on a polysaccharide molecular structure were no longer saturated by mAb binding at these concentrations (Fig. 6b). Furthermore, the binding reaction was probed at different time points (0.5–5 h), to establish a robust assay window where results would not be impacted by the binding kinetics. Assay results are very consistent between one to five hours of binding time (Supplementary Table 2 and Supplementary Fig. 2).
a Chromatogram overlay of antibody–polysaccharide complex (APC) formed under five different Ps concentrations. APC is eluted out at ~8 min and is separated from unbound antibody (eluted at ~9.8 min) (b). A 7-point polysaccharide antibody titration curve. With same antibody concentration at each point, a linear response was observed when polysaccharide concentration ([Ps]) is ≤0.15 µg/mL. The linear line started to bend when polysaccharide concentration increased to above 0.15 µg/mL, where antibody to polysaccharide ratio dropped below what is required to saturate all binding epitopes on the polysaccharide.
Initial analytical performance assessment
After established proof-of-concept on ST4, the assay was expanded to the remaining PCV15 serotypes for free polysaccharide (free Ps) quantitation. Initial assessments of assay performance parameters required by FDA and ICH for method validation41,42 were performed in the following sections. These parameters include assay specificity, linearity, range, precision and accuracy as discussed in the following paragraphs in this section.
The serotype (ST) knockout (KO) sample (“Methods”) for each of the PCV15 serotypes was prepared to demonstrate assay specificity. Antibody–polysaccharide complex (APC) was only observed in the binding reaction of an anti-ST monoclonal antibody (mAb) with the Ps of the same serotype. There is no APC observed in the reaction between an anti-ST mAb and the corresponding serotype KO sample. (Supplementary Fig. 3). This suggests that APC arises from the serotype-specific binding between the anti-ST mAb and Ps of the same serotype.
An anti-ST mAb was bound to the corresponding serotype standard at concentrations from low to high to generate a 5-point standard curve for each serotype. The standard curve was demonstrated by APC fluorescence signal peak areas vs the polysaccharide concentrations in these binding reactions. Excellent linearity (R2 > 0.99) has been observed for fourteen of the 15 serotypes, good linearity (R2 > 0.98) has been observed for ST33F. The standard curve range is from 0.01 to 0.15 µg/mL for most serotypes, or lower for some serotypes (Table 2 and Supplementary Fig. 4). This demonstrates that the assay is capable of serotype-specific quantitation of polysaccharide at sub-µg/mL levels.
Precision for this assay was evaluated by three experiments on two instrument systems. Before binding to a mAb, the PCV15 vaccines were prepared by an affinity capture step to remove CRM197 conjugate species. The resulting vaccine sample containing only free polysaccharide was bound to each of the fifteen anti-ST mAb individually in a binding reaction that mimicked the polysaccharide standard. The vaccine binding reactions were analyzed on SEC HPLC, and the APC peak for each ST was compared with the corresponding standard curve to calculate the free Ps in the vaccine (Supplementary Fig. 5). The precision for all 15 serotypes is summarized in Table 3. The serotype-specific chromatography assay has precisions better than 13.3% CV(coefficient of variation) for all serotypes.
Assay accuracy was evaluated with six representative serotypes. The standard curve has a range of 0.01–0.15 µg/mL for these serotypes. The middle of the highest standard concentration (0.075 µg/mL) is considered as the nominal assay concentration, and one-fifth of the nominal concentration (0.015 µg/mL) as lower limit of quantitation (LLOQ). The assay accuracy was assessed by the spike recoveries at the nominal concentration and at LLOQ. For all tested STs, excellent spike recoveries (97–103%) were observed for the spikes at nominal concentration. Spike recoveries at LLOQ were also acceptable42 at 83–119% (Table 4 and Supplementary Table 3). These demonstrate good assay accuracy. The free polysaccharide from the antibody chromatography assay are generally in agreement with results from our in-house sandwich ELISA assay and are in line with free Ps input expected from the vaccine formulation (Supplementary Table 4).
Multiplex serotype-specific chromatographic assay for conjugate quantitation
With the serotype-specific free Ps assay in hand, we sought to expand the assay to include conjugated polysaccharides. The challenge for a conjugate assay derives from the fact that protein signal detected from a serotype-specific APC complex not only represents mAbs binding to polysaccharides, but also CRM197 carrier proteins already on the conjugate. To mask out the background signal from the carrier protein, Alexa Fluor (AF) fluorescent (FLR) tag-labeled anti-ST mAb was generated and the fluorescence-tagged mAb was used for binding. The resulting fluorescence-labeled APC was detected by the FLR tag signal in a channel distinctive from that used for native protein detection (Ex/Em 280/352 nm). This FLR-labeled antibody chromatography assay can also be adapted on a multiplex format, since HPLC detectors have the ability to do multi-channel FLR or UV detection simultaneously. With this in mind, we labeled four anti-ST mAbs, each with a unique FLR tag (anti- ST4-AF350, ST5-AF430, ST6A-AF555, and ST14-AF633, “Methods”) (Supplementary Fig. 6). Four distinctive fluorescence channels were set in a single HPLC method, each is corresponding to one of the FLR tags on a mAb. In this way, one serotype can only be detected on the desired FLR channel, with no signal observed on the other three channels (Supplementary Fig. 7). Coupled with antibody’s binding specificity, a multiplex serotype-specific assay for PCV total Ps (conjugated + free Ps) has been established. It is worth to note that this multiplex assay can be applied to both conjugated and unconjugated polysaccharides.
A PCV vaccine standard with conjugated ST4, ST5, ST6A, and ST14 were used to generate four standard curves in a single HPLC run. All four conjugate serotypes showed good linearity in the respective standard curve (R2 > 0.99) (Fig. 7 and Supplementary Table 5). A 24-valent conjugate vaccine (PCV24)43 with a target formulation total Ps (conjugate Ps + free Ps) concentration of 8 µg/mL for each serotype was analyzed in a single HPLC run. The measured Ps concentrations are within ±11% of the target potency value (Table 5 and Supplementary Table 6) for all four tested serotypes (ST4, ST5, ST6A, and ST14), complying with the normal acceptance criteria of ±30% of the label claim for polysaccharide conjugated vaccines23,25. The 4-in-1 multiplex assay format reduces the HPLC runs needed to fully quantify all serotypes in the PCV24 from 24 to 6.
Discussion
Pneumococcal disease is a major cause that leads to serious medical conditions. Vaccine development for this disease can be greatly benefited from a simple and robust serotype-specific antigen assay for vaccine products. Previous reports of such assay for pneumococcal conjugate vaccines are scarce and lack of recent advance. The only literature-reported method is a nephelometry method. The nephelometry method uses light scattering to detect the antibody–polysaccharide complex formed in the solution. The nephelometry observed 82–119% variation from specification. The linearity range for this method is rather narrow due to the general non-linear nature of direct light scattering response to concentrations. Also, since the assay is performed in a direct batch flow-through mode without separation of or washing off sample matrix, sample formulation matrix could potentially impact the quality of the results. For certain conjugate sample, treatment by trypsin may be required to achieve optimal result in nephelometry25. In the antibody-assisted HPLC assay, antibody–polysaccharide complex is separated from sample matrix on chromatography. Therefore, matrix interference can be minimized. The only sample preparation procedure is the dilution with sample or binding buffer before analysis. Generally, a 10-fold to 15-fold linearity range can be easily established on an antibody HPLC assay.
Presumably, another preferred assay platform for such serotype-specific quantitation would be the sandwich ELISA assay, although there is no literature report about its application on a pneumococcal vaccine. For each serotype, sandwich ELISA assay platform would require three antibodies: a capture antibody, a detection antibody and an enzyme-linked antibody for the chemiluminescence signal. Several concentration points would be required for generating a titration curve in ELISA analysis. ELISA sample and plate preparations involve relatively manual and lengthy processes. In antibody-assisted HPLC, sample preparation and antibody binding are relatively simple and straightforward. The HPLC analysis is automatic and can be easily transferred between labs without much variation. Antibody-assisted HPLC only requires one antibody for each serotype in comparison with three for the sandwich ELISA. This will reduce some burden on critical reagent generation and management. Even though antibody-assisted HPLC is benefit from signal enhancement from binding multiple antibodies to one polysaccharide, generally it is still less sensitive than an ELISA assay. ELISA could analyze samples at ng/mL concentration, whereas antibody HPLC worked best at ~0.005 to 0.20 µg/mL range in our experience. All assays discussed above could share the same control strategies for reference standards and antipolysaccharide antibody reagents.
Good linearity (generally R2 ≥ 0.99) has been achieved for the antibody-enhanced HPLC assay. For residual free polysaccharide analysis in the PCV15 vaccine, we have reported precision of %CV from 0.7% to 13.3% across all 15 serotypes. In our matrix based approach to assess assay accuracy, a 97–103% recovery was obtained for samples tested at nominal concentration, with six different serotypes were analyzed. The assay performance meets the regulatory requirements for initial validation assessment of both chromatographic assays (CCs) and ligand binding assays (LBAs)42. Since each free polysaccharide serotype existed in different concentrations, these indicated that analytes with varied concentrations performed similarly well in the antibody HPLC assay. If a formal method validation with more sample batches and data points is successful in the future, the assay could be used for commercial product release or quality control. We also established proof-of-concept for using fluorescent tag-labeled antibodies to quantify polysaccharide conjugates on a multiplex HPLC assay platform. A 5–11% variation from product specification was observed for the four tested serotypes. This is well below regulatory criteria of ±30% of specification, for a vaccine content/potency release assay23,25. A fourfold improvement on assay throughput would greatly reduce resource needed for analysis of a multi-valent vaccine containing 20 plus serotypes. Further development work is needed to assess the full spectrum of the assay performance on this novel assay format. An antibody-assisted HPLC assay can be easily developed to analyze other polysaccharide and vaccine products, when quality of antibody is reliable and under good control.
Although the antibody HPLC assay was only established on the size-exclusion chromatography (SEC) platform in this report, conceivably it can also be established with other chromatographic separation methods, such as ion-exchange (IEX) or hydrophobic interaction chromatography (HIC) or with electrophoresis separation methods, such as capillary zone electrophoresis (CZE) or capillary isoelectric focusing (cIEF). Furthermore, the antibody-enhanced chromatography assay leveraged on the structure features of antibody binding to a multi-epitope antigen (polysaccharide). This structure-guided assay development approach can be applied and explored for future assay developments. After the assay is established, in turn it can be used to study antibody–polysaccharide binding affinity and kinetics, characterize the antibody–polysaccharide complex and assess quantity and quality of serum antibody obtained from clinical trial subjects or patients.
In this study, a novel antibody-enhanced chromatography assay for serotype-specific quantitation of polysaccharide was developed for a pneumococcal conjugate vaccine. This represents the first time report of a serotype-specific antigen chromatography assay for a multi-valent vaccine. Good assay performance (specificity, linearity, precision and accuracy) has been demonstrated for the quantitation of free polysaccharides in the vaccine. Proof-of-concept was demonstrated on a multiplex HPLC assay platform, when fluorescent tag-labeled antibodies were introduced. This multiplex platform is capable of quantifying both protein-conjugated and unconjugated polysaccharides in a multi-valent vaccine. This simple and automatic assay platform should offer great impact and broad applications in the research and development of vaccines and biologics.
Methods
Reagents and materials
Bis-Tris-HCl 1 M solution was purchased from Rigaku Reagents, Inc. (Seattle, WA). Sodium chloride 5 M solution was bought from Promega Corporation (Madison, WI). Pierce™ Protein A/G Magnetic Beads, Alexa Fluor™ NHS Esters and Zeba Spin desalting columns were purchased from Thermo Fisher. PBS buffer was purchased from Gibco. Antibody-binding buffer was made by our in-house reagent group.
MAbs and FLR tag-labeled mAbs
Pneumococcal anti-ST mAbs and anti-CRM197 mAbs were generated and provided to authors by our in-house antibody generation and critical reagent teams44,45. All mAbs were generated in-house for this research. All mAb clones were generated from our internal research teams, except clones for anti-serotypes 1 and 19F mAbs. Anti-serotypes 1 and 19F mAbs were licensed from the University of Alabama at Birmingham (UAB). The mAb species and source were listed in Supplementary Table 7. Each of the anti-polysaccharide mAb was diluted from the stock mAb solution as in Supplementary Table 8, before added into a binding reaction.
FLR tag-labeled anti-ST mAbs were prepared by conjugation to corresponding Alexa Fluor™ NHS esters as exemplified by labeling of anti-ST4 with Alexa Fluor™ 350 (AF350) below.
An anti-ST4 mAb was incubated with excess equivalents of Alexa Fluor™ 350 NHS ester PBS buffer for three hours at ambient temperature. The reaction was purified by desalting through a Zeba Spin desalting column. AF350-labeled anti-ST4 (anti-ST4-AF350) mAb was collected from the column. In the same fashion, anti-ST5, anti-ST6A and anti-ST14 mAbs were prepared as anti-ST5-AF430, anti-ST6A-AF555 and anti-ST14-AF633, respectively (with Alexa Fluor™ 430, Alexa Fluor™ 555 and Alexa Fluor™ 633).
The four FLR-labeled anti-ST mAbs are mixed together as a cocktail (FLR-mAbs) with 0.2 mg/mL of each mAb and used to multiplex with a multivalent PCV vaccine.
PCV vaccine product and PCV conjugate and free polysaccharide standards
PCV-free polysaccharide standards, conjugated polysaccharide standards, and research batches of PCV vaccine were generated and provided to authors by our vaccine process and formulation groups46.
Serotype-specific knockout (KO) polysaccharide (Ps) samples are prepared by authors by mixing fourteen out of the fifteen PCV15 Ps standards together with the target Ps serotype missing (knockout).
Preparation of PCV vaccine sample for free Ps analysis
The CRM197-conjugated Ps species were removed from the vaccine product by an affinity capture step. In short, CRM197 conjugated Ps were removed from the vaccine solution by immunoprecipitation with Pierce™ Protein A/G magnetic beads coated with anti-CRM197 monoclonal antibodies (mAbs) at 20 µg/mL each. After removal of the magnetic beads from the solution, the supernatant contained only unconjugated free Ps and was used to bind each anti-ST mAb in the antibody-binding reactions and analyzed on SEC.
Preparation of anti-ST mAb binding reactions for free Ps analysis
The PCV Ps standards were bound to the corresponding anti-ST mAb in the same or similar fashion that described in Supplementary Table 9a (Standard Binding Table) to generate a serotype-specific standard curve for that type. The reactions are incubated at ambient temperature for one or two hours before injected on HPLC. Each Ps serotype has its own standard curve.
The vaccine solution prepared from immunoprecipitation with anti-CRM197 mAbs was used to bind each anti-ST mAb as in Supplementary Table 9b. Each vaccine preparation will bind to all 15 anti-ST mAbs individually. The reactions are incubated at ambient temperature for the same time period as the Ps standards, before injected on HPLC.
Multiplex binding reactions for PCV total Ps (conjugated Ps + free Ps) analysis
A mixed PCV standard that contains 1 µg/mL each of ST4, ST5, ST6A, and ST14 total Ps (conjugated Ps + free Ps) was provided by our vaccine formulation group. The PCV24 vaccine53 was also provided by our vaccine formulation group.
The PCV standard was multiplexed with the FLR-labeled mAb cocktail (FLR-mAbs) as in Supplementary Table 10 and incubated at ambient temperature for two hours before HPLC analysis. The PCV24 vaccine was multiplexed with FLR-mAbs in the same fashion as in Supplementary Table 11 and incubated for the same time period as the standards before analysis.
Size-exclusion chromatography (SEC) conditions
The chromatography system was set either on a Waters Alliance (Waters Corporation, Milford, MA), or an Agilent 1260 Infinity II Bioinert system (Agilent, DE, USA), equipped with a quaternary pump, sample manager, column compartment, fluorescence (FLR) and photodiode array (PDA or DAD) detectors. The FLR was detected using excitation (Ex) wavelength at 280 nm and emission (Em) wavelength at 352 nm (Ex/Em 280 nm/352 nm). UV absorbance was recorded at 280 nm (A280).
For detection involving FLR-labeled mAbs, the FLR detections were set at Ex/Em wavelengths corresponding to the FLR tag on the mAbs. Four FLR detection channels were set on the Waters Alliance HPLC system in a single method to allow simultaneous detections of signal carried from four labeled mAbs. The anti-ST4-AF350 was detected at Ex/Em of 346 nm/442 nm, anti-ST5-AF430 at 433 nm/541 nm, anti-ST6A-AF555 at 555 nm/565 nm, anti-ST14-AF633 at 633 nm/647 nm.
General chromatography conditions are listed in Supplementary Table 12 (Chromatography Condition Table). Either Chromatography Condition-A or Chromatography Condition-B was used for a certain serotype.
SEC-MALS-UV-RI analysis of antibody–polysaccharide complex (APC)
The SEC-MALS-UV-RI analysis was performed in the method and chromatography system below. The chromatography condition was the same to the Chromatography condition-A in Supplementary Table 12, except a Tosoh TSKgel G4000PWxL column (7.8 × 300 mm) was used. A multi-angle light scattering (MALS) detector (MiniDAWN®) and a refractive index (RI) detector (Optilab® T-rEX) (Wyatt Technology Corp., Santa Barbara) were added to the system and connected in tandem with the UV detector. All data were collected and analyzed with ASTRA 7 software. A first-order fit Zimm formalism with Astra protein conjugate method was used for Mw calculations47,48,49.
Anti-ST1 mAb was complexed with ST1 polysaccharide (Ps) standard at different mAb/Ps ratio as in Supplementary Table 13. The binding reactions were incubated at ambient temperature for 2 h and injected on the SEC-MALS-UV-RI system for analysis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Geno, K. A. et al. Pneumococcal capsules and their types: past, present, and future. Clin. Microbiol. Rev. 28, 871–899 (2015).
Daniels, C. C., Rogers, P. D. & Shelton, C. M. A review of pneumococcal vaccines: current polysaccharide vaccine recommendations and future protein antigens. J. Pediatr. Pharm. Ther. 21, 27–35 (2016).
Musher, D. M. et al. Natural and vaccine-related immunity to Streptococcus pneumoniae. J. Infect. Dis. 154, 245–256 (1986).
Weller, S., Reynaud, C. A. & Weill, J. C. Vaccination against encapsulated bacteria in humans: paradoxes. Trends Immunol. 26, 85–89 (2005).
Butler, J. C. et al. Pneumococcal polysaccharide vaccine efficacy: an evaluation of current recommendations. J. Am. Med. Assoc. 270, 1826–1831 (1993).
Niedermana, M. S. et al. Efficacy and effectiveness of a 23-valent polysaccharide vaccine against invasive and noninvasive pneumococcal disease and related outcomes: a review of available evidence. Expert Rev. Vaccines 20, 243–256 (2021).
Gruber, W. C., Scott, D. A. & Emini, E. Development and clinical evaluation of Prevnar 13, a 13-valent pneumococcal CRM197 conjugate vaccine. Ann. NY Acad. Sci. 1263, 15–26 (2012).
Cannon, K. et al. A trial to evaluate the safety and immunogenicity of a 20-valent pneumococcal conjugate vaccine in populations of adults ≥65 years of age with different prior pneumococcal vaccination. Vaccine 39, 7494–7502 (2021).
Prymula, R. & Schuerman, L. 10-valent pneumococcal nontypeable Haemophilus influenzae PD conjugate vaccine: synflorix. Expert Rev. Vaccines 8, 1479–1500 (2009).
Greenberg, D. et al. Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine (PCV15) in healthy infants. Vaccine 36, 6883–6891 (2018).
Platt, H. L. et al. A phase 3 trial of safety, tolerability, and immunogenicity of V114, 15-valent pneumococcal conjugate vaccine, compared with 13-valent pneumococcal conjugate vaccine in adults 50 years of age and older (PNEU-AGE). Vaccine 40, 162–172 (2022).
Peterson, J. T. et al. Safety and immunogenicity of 15-valent pneumococcal conjugate vaccine compared to 13-valent pneumococcal conjugate vaccine in adults ≥65 years of age previously vaccinated with 23-valent pneumococcal polysaccharide vaccine. Hum. Vaccin Immunother. 15, 540–548 (2019).
Lexau, C. A. et al. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. J. Am. Med. Assoc. 294, 2043–2051 (2005).
Isaacman, D. J., McIntosh, E. D. & Reinert, R. R. Burden of invasive pneumococcal disease and serotype distribution among Streptococcus pneumoniae isolates in young children in Europe: impact of the 7-valent pneumococcal conjugate vaccine and considerations for future conjugate vaccines. Int J. Infect. Dis. 14, e197–e209 (2010).
Masomian, M., Ahmad, Z., Gew, L. T. & Poh, C. L. Development of next generation Streptococcus pneumonia vaccines conferring broad protection. Vaccines 8, 132 (2020).
Poolman, J. T. Expanding the role of bacterial vaccines into life-course vaccination strategies and prevention of antimicrobial-resistant infections. npj Vaccines 5, 84 (2020).
Frasch, C. E. Preparation of bacterial polysaccharide–protein conjugates: analytical and manufacturing challenges. Vaccine 27, 6468–6470 (2009).
Stark, J. C. et al. On-demand biomanufacturing of protective conjugate vaccines. Sci. Adv. 7, eabe9444 (2021).
Biemans, R., Micoli, F. & Romano, M. R. Glycoconjugate vaccines, production and characterization. Recent Trends Carbohydr. Chem. 2, 285–313 (2020).
Verch, T., Trausch, J. J. & Shank-Retzlaff, M. Principles of vaccine potency assays. Bioanalysis 10, 163–180 (2018).
Manceur, A. P. & Kamen, A. A. Critical review of current and emerging quantification methods for the development of influenza vaccine candidates. Vaccine 33, 5913–5919 (2015).
Byrne-Nash, R. T. et al. VaxArray potency assay for rapid assessment of “pandemic” influenza vaccines. npj Vaccines 3, 1–11 (2018).
Phugare, S. et al. Quantitation of novel pentavalent meningococcal polysaccharide conjugate vaccine (Men A-TT, Men C-CRM, Men Y-CRM, Men W-CRM, Men X-TT) using sandwich ELISA. Vaccine 38, 7815–7824 (2020).
Gillis, J. H. et al. Multiplexed VaxArray immunoassay for rapid antigen quantification in measles and rubella vaccine manufacturing. Vaccine 9, 100–113 (2021).
Lee, C. Quality control of polyvalent pneumococcal polysaccharide-protein conjugate vaccine by nephelometry. Biologicals 30, 97–103 (2002).
Sanyal, G., Särnefält, A. & Kumar, A. Considerations for bioanalytical characterization and batch release of COVID-19 vaccines. npj Vaccines 6, 53 (2021).
Deng, J. Z. et al. RP-UPLC method for oncolytic coxsackievirus viral protein separation and empty to full capsid quantification. Hum. Gene Ther. https://doi.org/10.1089/hum.2022.013 (2022).
Lei, Q. P., Lamb, D. H., Heller, R. & Pietrobon, P. Quantitation of low level unconjugated polysaccharide in tetanus toxoid-conjugate vaccine by HPAEC/PAD following rapid separation by deoxycholate/HCl. J. Pharm. Biomed. Anal. 21, 1087–1091 (2000).
Gao, F., Lockyer, K., Burkin, K., Crane, D. T. & Bolgiano, B. A physico-chemical assessment of the thermal stability of pneumococcal conjugate vaccine components. Hum. Vaccines Immunotherapeutics 10, 2744 (2014).
Hadidi, M., Buckley, J. J. & Zydney, A. L. Effects of solution conditions on characteristics and size exclusion chromatography of pneumococcal polysaccharides and conjugate vaccines. Carbohydr. Polym. 152, 12–18 (2016).
Abdelhameed, A. S. et al. Solution conformation and flexibility of capsular polysaccharides from Neisseria meningitidis and glycoconjugates with the tetanus toxoid protein. Sci. Rep. 6, 35588 (2016).
Anish, C., Beurret, M. & Poolman, J. Combined effects of glycan chain length and linkage type on the immunogenicity of glycoconjugate vaccines. npj Vaccines 6, 150 (2021).
Wessels, M. R. & Kasper, D. L. Antibody recognition of the type 14 pneumococcal capsule: evidence for a conformational epitope in a neutral polysaccharide. J. Exp. Med. 169, 2121–2131 (1989).
Zou, W & Jennings, H. J. The conformational epitope of type III group B streptococcus capsular polysaccharide. Adv. Exp. Med. Biol. 491, 473–484 (2001).
Vulliez-Le Normand, B. et al. Structures of synthetic O-antigen fragments from serotype 2a Shigella flexneri in complex with a protective monoclonal antibody. PNAS USA 105, 9976–9981 (2008).
Soliman, C., Pier, G. B. & Ramsland, P. A. Antibody recognition of bacterial surfaces and extracellular polysaccharides. Curr. Opin. Struct. Biol. 62, 48–55 (2020).
Henriques, P. et al. Structure of a protective epitope reveals the importance of acetylation of Neisseria meningitidis serogroup A capsular polysaccharide. PNAS USA 117, 29795–29802 (2020).
Deng, J. Z. et al. SEC coupled with in-line multiple detectors for the characterization of an oncolytic coxsackievirus. Mol. Ther.: Oncolytics 24, 139–147 (2022).
Deng, J. Z. et al. Multi-attribute characterization of pneumococcal conjugate vaccine by size-exclusion chromatography coupled with UV-MALS-RI detections. Vaccine 40, 1464–1471 (2022).
Deng, J. Z., Lin, J., Chen, M., Lancaster, C. & Zhuang, P. Characterization of high molecular weight pneumococcal conjugate by SEC-MALS and AF4-MALS. Polymers 14, 3769 (2022).
International Conference on Harmonisation. Validation of Analytical Procedures: Text And Methodology Q2 (R1). (International Conference on Harmonisation, 2005).
Center for Drug Evaluation and Research (CDER) and Center for Veterinary Medicine (CVM). Food and Drug Administration Bioanalytical Method Validation: Guidance for Industry https://www.fda.gov/files/drugs/published/Bioanalytical-Method-Validation-Guidance-for-Industry.pdf (2018).
McGuinness, D. et al. Immunogenicity of PCV24, an expanded pneumococcal conjugate vaccine, in adult monkeys and protection in mice. Vaccine 39, 4231–4237 (2021).
Chen, Z. et al. Human monoclonal antibodies isolated from a primary pneumococcal conjugate vaccine demonstrates the expansion of an antigen-driven hypermutated memory B cell response. BMC Infect. Dis. 18, 613 (2018).
Cox, K. S. et al. Efficient isolation of monoclonal antibodies from antigen-specific human plasmablasts and memory B cells from a pneumococcal conjugate vaccine. J. Immunol. 200, 180 (2018).
Ahl, P. L. et al. Methods for producing Streptococcus pneumoniae capsular polysaccharide carrier protein conjugates. Patent application US20210252125A1.
Wyatt, P. J. Light scattering and the absolute characterization of macromolecules. Anal. Chim. Acta 272, 1–40 (1993).
Zimm, B. H. The scattering of light and the radial distribution function of high polymer solutions. J. Chem. Phys. 16, 1093–1099 (1948).
Kendrick, B. S., Kerwin, B. A., Chang, B. S. & Philo, J. S. Online size-exclusion high-performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein-protein or protein-ligand association states. Anal. Biochem. 299, 136–146 (2001).
Acknowledgements
Funding of this work was provided entirely by Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA. We would like to thank our antibody generation and critical reagent groups for providing antibodies used in this study. We would also thank the University of Alabama at Birmingham (UAB) for providing us the antibody clones for serotypes 1 and 19F. We also would like to extend our gratitude to colleagues in our Vaccine Process and Formulation Development teams for providing us with purified PCV vaccine materials and standards. We would like to thank Drs. Richard Rustandi, Zhifeng Chen, Brittany Paporello, and Caroline McGregor for reviewing this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: J.Z.D.; Ab-HPLC method development and data collection: J.Z.D., M.L., N.K., and A.D.; ELISA and affinity capture: F.A., M.G., and R.S.; writing-original draft, literature review and data interpretation: J.Z.D.; writing-review and editing: P.Z., A.D., M.L., and M.G.; supervision and program administration: J.Z.D. and P.Z.; all authors have reviewed the manuscript and agreed to the contents and author contributions.
Corresponding author
Ethics declarations
Competing interests
All authors are current employees of Merck & Co., Inc., Rahway, NJ, USA, and may hold stocks or stock options of this company. The authors declare that none of the authors has competing financial interests that can affect the data collection and data interpretation.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Deng, J.Z., Kuster, N., Drumheller, A. et al. Antibody enhanced HPLC for serotype-specific quantitation of polysaccharides in pneumococcal conjugate vaccine. npj Vaccines 8, 2 (2023). https://doi.org/10.1038/s41541-022-00584-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41541-022-00584-9
This article is cited by
-
Serotype-specific quantification of residual free polysaccharide in multivalent pneumococcal conjugate vaccines
Glycoconjugate Journal (2024)
-
Characterization of pneumococcal serotype 7F in vaccine conjugation
Glycoconjugate Journal (2023)