Abstract
The rVSV-ZEBOV Ebolavirus vaccine confers protection within days after immunization, suggesting the contribution of innate immune responses. We report modulation of rVSV-ZEBOV vaccinee blood CD56+ NK cell numbers, NKG2D or NKp30 surface receptor expression, Killer Immunoglobulin-like Receptor (KIR)+ cell percentages and NK-cell-related genes on day 1 post immunization. Inverse correlations existed between the concentration of several plasma cytokines and inhibitory KIR+ CD56dim or cytokine-responsive CD56bright NK cells. Thus, NK cells may contribute to the early protective efficacy of rVSV-ZEBOV in humans.
Similar content being viewed by others
Introduction
Ebolavirus continues to cause deadly epidemics. The live-attenuated rVSV-ZEBOV vectored vaccine has now been employed in over 255,000 people in “ring vaccination” trials in Africa1,2, mediating 97.5% protection if administered at least 10 days prior to contact with infected individuals, as well as partial protection for shorter periods between vaccination and exposure (https://www.who.int/csr/resources/publications/ebola/ebola-ring-vaccination-results-12-april-2019.pdf). Early protection is vital to interrupt viral outbreaks. As 2 weeks are required for ZEBOV glycoprotein (GP)-specific IgG antibodies to appear in the blood of most vaccinees3, and at least 3–7 days for IgM4, innate vaccine-elicited responses may contribute to the rapid protection.
NK cells have clear roles in antiviral responses5. Cytometric gating of CD56+ CD3− cells allows relatively specific identification of circulating blood NK cells. The intensity of CD56 reflects cell functionality with CD56bright NK cells showing superior responses to soluble factors whereas CD56dim NK cells are geared towards recognizing cell surface-bound factors6. NK cell function is governed by a large number of activating and inhibitory signals. For example, NKG2D is a surface activating receptor on NK and T cells that recognizes pathogen-associated molecular patterns, whereas Killer Immunoglobulin-like Receptors (KIR) recognize host HLA molecules, and can represent major inhibitory receptors5,7 depending on immune education events with self-HLA ligands.
Many cytokines and chemokines detected at elevated levels immediately after rVSV-ZEBOV immunization (IL-1ra, IL-6, IL-10, TNF-α, MCP-1/CCL2, and MIP-1β/CCL4)8,9 can also influence the maturation/trafficking of NK cells and may be secreted by them5,10,11,12. Early rVSV-ZEBOV responses (including IL-15 and type I interferon) were hypothesized to activate NK cells4, and additionally, certain post-vaccination NK cell phenotypes have been shown to correlate with long-term vaccine-specific Ab responses8. This latter study focused on how very early immune responses to rVSV-ZEBOV could shape immune memory, whereas potential augmentation of immediate antiviral protection via vaccine-induced innate immunity was not evaluated. Therefore, to explore the potential contribution of NK cells to early protection from Ebolavirus, we investigated the changes in NK cell numbers, phenotypes and modulated gene-transcripts in a subset (n = 22) of Geneva vaccinees (NCT02287480)3,13, after an rVSV-ZEBOV high dose (HD)−i.e. currently used in Africa, compared to a low dose (LD), occurring in the first week after vaccination.
Results
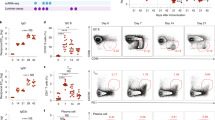
Repeated blood draws (Fig. 1a) were used to monitor the proportions and phenotype of major NK cell subsets (Fig. 1b) in fresh PBMCs following rVSV-ZEBOV administration. Baseline variability of absolute numbers and percentages of NK cells (Supplementary Fig. 1) were observed in the cohort; hence, we calculated the ratios of any changes in NK cells at the intra-participant level. On day 1, HD vaccination induced pronounced and transient reductions in total NK cell counts and percentages (Fig. 1c and Supplementary Fig. 1), including both CD56bright and CD56dim subpopulations (Fig. 1d, e). In contrast, LD vaccination only significantly reduced CD56bright NK cell numbers (Fig. 1d), which represent a minority of circulating NK cells. A rebound or expansion was seen after HD/LD vaccination, with higher numbers (Fig. 1c−e) and percentages (Supplementary Fig. 1) of NK cells on days 3 or 7 compared to baseline. Unsupervised blood transcription module (BTM) analysis of HD vaccinees identified gene module LI.M61.0 “enriched in NK cells (II)”, as the most downregulated of all 346 BTMs on day 1 (manuscript in press), returning to baseline levels by day 3 (Supplementary Fig. 1). Many NK-cell-related genes from this BTM, including indicators of activation (Klrk1; encoding NKG2D), potential inhibition (Kir2DL3), maturation (Nkp80), migration (S1pr5) and cytokine responsiveness (Tgfbr3; encoding IL2RB), were downregulated on day 1 compared to day 0 (Fig. 1f). Therefore, cytometry and transcriptomics suggested early and transient total NK cell reductions in the blood after HD vaccination.
a Study summary of volunteers injected with high (HD) or low dose (LD) of rVSV-ZEBOV, or placebo (P). b Dot plots depict representative HD vaccinee PBMCs at baseline, gate values indicate cohort-wide % of pre-gated singlet, non-debris, leukocytes ± SD, followed by positive selection of either total leukocytes (dark blue gate), or non-granulocyte “lymphocyte”-like (light blue gate) followed by CD56bright or CD56dim NK cell selection. c−e Absolute number of c total NK cells (combined CD56bright and CD56dim NK cells), d CD56bright NK and e CD56dim NK cells. Violin plot symbols (open circles) show the intra-vaccinee net ratio compared to the baseline values (dashed lines), as well as median (thick red line), and interquartile (thin red line) values, shaded vaccine doses; (black squares) HD, (gray squares) LD, (open squares) placebo, and enclosed shaded horizontal width indicates the probability of obtaining the corresponding y-axis value. A one sample t test compared days 1, 3, or 7 to baseline ratio values with an expected value of 1. f Targeted sequencing was used to determine the average day 1, 3, and 7 to baseline fold change for genes in BTM LI.M61.0 “enriched in NK cells (II)” in HD vaccinees, indicated by the color scale, and compared using the edgeR glmtreat command with Benjamini−Hochberg multiple testing corrections. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Phenotypic changes in NK cells subsets were investigated, with all significantly modulated markers shown in Fig. 2a and Supplementary Fig. 1, and nonmodulated markers listed in the Methods. A high vaccine dose increased the expression of two activation markers; NKG2D (Fig. 2b) and CD56 (Supplementary Fig. 1) in CD56dim NK cells, and reduced NKp30 expression in both CD56bright and CD56dim subsets (Fig. 2c, Supplementary Fig. 1). The proportion of KIR2DL1+ CD56dim NK cells was significantly reduced in HD and LD vaccinees (Fig. 2d), as well as the KIR3DL1+KIR3DS1+ (CD158e1/e2+) subset after HD vaccination (Fig. 2e). Most phenotypic changes returned to baseline levels by days 3 or 7 (Fig. 2b−e), during the re-expansion of total CD56+ cell numbers and percentages (Fig. 1c, Supplementary Fig. 1). The few changes persisting beyond day 1 included NKG2D downregulation in HD vaccinees (Fig. 2b, Supplementary Fig. 1), and increased NKp30 expression in CD56dim NK cells after LD vaccination (Fig. 2c). Overall, these phenotypic changes indicate the significant activation of CD56dim NK cells in HD, and to a lesser extent in LD rVSV-ZEBOV vaccinees.
a Cytometry contour plots of the indicated cell subsets, colored as indicated, from a representative HD vaccinee at baseline showing vaccine-modulated surface markers after gating as described in Fig. 1a. The indicated KIR+ subset gates are shown as open rectangles with values of cohort-wide % of total NK cells ± SD, and control cells represent total leukocytes from unstained samples. b−e Fold change analysis of CD56dim NK cells compared to the baseline, depicted and compared with statistics as described for the violin plots in Fig. 1, using the geometric MFI of surface markers b NKG2D, c NKp30, or percentages of d KIR2DL1+ or e KIR3DL1/S1+ cells within the CD56dim NK cell gate. f Two-tailed Pearson analysis comparing the indicated NK cell phenotypic parameters with plasma cytokine concentrations from matched vaccinees, all on day 1 post vaccination. Pearson r values were calculated using a 95% CI. *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether vaccine-modulated NK-cell-related parameters were related to each other, to previously identified individual cytokine levels9 or to our previously described rVSV-ZEBOV plasma cytokine signature9, we performed Pearson comparisons between pairs of parameters from day 1 post immunization, including dose-based data stratification (HD, left panel; LD, right panel; Fig. 2f, Pearson r and P values in Supplementary Tables 1–4). The plasma signature represents a combined score of 6 cytokines/chemokines identified using the entire Geneva cohort9. A greater number of significant correlations existed in HD than LD vaccinees, again emphasizing the dose-related effects of rVSV-ZEBOV. In HD vaccinees, negative correlations existed between the proportion of KIR2DL1 receptor-expressing CD56dim NK cells and activating NKG2D receptor expression levels in CD56bright NK cells, and also with levels of NK activating IL-1ra10 or TNF-α12 plasma levels (left panel, Fig. 2f). Strikingly, the plasma level of three cytokines, IL-6, MCP-1, and MIP-1β, were inversely correlated with the number of circulating CD56bright “cytokine-responsive” NK cells. A nonsignificant negative correlation trend existed between the plasma cytokine signature in HD vaccinees and the numbers of cytokine-responsive CD56bright NK cells (r = −0.66, P = 0.076; left panel, Fig. 2g), suggesting enhanced CD56bright NK cell efflux from the blood in vaccinees with higher combined plasma cytokine concentrations.
Discussion
This study demonstrates that the dose of rVSV-ZEBOV impacts circulating NK cell responses because heightened activation (increased NKG2D5 and CD566), depletion of NK cells and many inverse correlations between plasma cytokine levels and percentages of potentially inhibitory KIR+ CD56dim or numbers of cytokine-responsive CD56bright NK cells were only detected after HD vaccination.
Circulating NK cells are likely to be activated by many of the cytokines induced by rVSV-ZEBOV4,8,9. Within the top 15 most upregulated BTMs, three were for cytokine families (manuscript in preparation) previously defined as strong NK cell activators, namely type I IFNs14, IL-1515, and TNF-α12. The striking NK cell depletion observed may be due to activation-induced cell death or potential margination, i.e. adherence of the activated cells to vessel walls. However, we postulate that rVSV-ZEBOV-activated NK cells emigrate from the blood towards vaccine replication sites, including injection sites where many innate cell subsets accumulate16. By day 3, the blood NK cell compartment was replenished with CD56+ cells expressing baseline levels of nearly all investigated markers except NKG2D, which could indicate an influx of NK cells of different developmental stages5 or VSV-mediated NKG2D modulation17. NKp30 modulation was also detected in our study, but this can lead to either IFN-γ or IL-10 induction, such that the consequence of this modulation cannot be inferred18. Together, these data led us to postulate that activated NK cell subsets in HD rVSV-ZEBOV vaccinees may contribute to the early control and clearance of Ebolavirus.
A limitation of our study is the small number of subjects that could be studied using fresh blood during a rapidly enrolling clinical study, and the absence of RNA analysis at the single-cell level. This latter omission precluded in-depth lineage analysis of CD56+ cells, and restricted the investigation of how changes in the proportions of other leukocyte subsets could affect bulk transcriptomic results. Nonetheless, the massive cellular efflux and elevated activation markers of CD56+ cells measured by flow cytometry provides a rationale to further decipher the role of NK cells after rVSV-ZEBOV vaccination.
Methods
Healthy adults (n = 115) participating in clinical trial NCT022874803 were injected with rVSV-ZEBOV HD (1 or 5 × 107 plaque forming units; pfu), LD (3 × 105 pfu) or saline placebo (P). The study was approved and overseen by the Ethics Commission of the Canton of Geneva, Switzerland; World Health Organization’s Ethics Review Committee, and complied with all relevant regulations for work with human participants. Written informed consent was obtained from all participants. Twenty-two participants were randomly selected for in-depth characterization of circulating CD56+ cells at the single-cell level by flow cytometry. When unblinded, these included 8, 11, and 3 participants in the HD, LD and placebo groups respectively. Calculation of absolute numbers, surface marker fluorescence intensity, and ratios are described in the Supplementary Methods.
Ion AmpliSeq™ Transcriptome Human Gene Expression Kits (Life Technologies) were used to sequence total RNA on an Ion Proton platform, and analyzed using the edgeR package. Blood draws, sample preparation, flow cytometry or transcriptomic analyses, and statistics are described in detail in the Supplementary Methods. Transcriptomic19 and flow cytometry data20 were uploaded to online databases.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data that support the findings of this study are available at https://doi.org/10.5281/zenodo.3415147 (transcriptomics)19, and https://doi.org/10.6084/m9.figshare.11886678 (flow cytometry and additional data)20.
Change history
12 February 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41541-020-00244-w
References
Gsell, P.-S. et al. Ring vaccination with rVSV-ZEBOV under expanded access in response to an outbreak of Ebola virus disease in Guinea, 2016: an operational and vaccine safety report. Lancet Infect. Dis. 17, 1276–1284 (2017).
Henao-Restrepo, A. M. et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet 386, 857–866 (2015).
Huttner, A. et al. The effect of dose on the safety and immunogenicity of the VSV Ebola candidate vaccine: a randomised double-blind, placebo-controlled phase 1/2 trial. Lancet Infect. Dis. 15, 1156–1166 (2015).
Marzi, A. et al. EBOLA VACCINE. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 349, 739–742 (2015).
Abel, A. M., Yang, C., Thakar, M. S. & Malarkannan, S. Natural killer cells: development, maturation, and clinical utilization. Front. Immunol. 9, 1–23 (2018).
Van Acker, H. H., Capsomidis, A., Smits, E. L. & Van Tendeloo, V. F. CD56 in the immune system: more than a marker for cytotoxicity? Front. Immunol. 8, 892 (2017).
Saunders, P. M. et al. A bird’s eye view of NK cell receptor interactions with their MHC class I ligands. Immunol. Rev. 267, 148–166 (2015).
Rechtien, A. et al. Systems vaccinology identifies an early innate immune signature as a correlate of antibody responses to the Ebola vaccine rVSV-ZEBOV. Cell Rep. 20, 2251–2261 (2017).
Huttner, A. et al. A dose-dependent plasma signature of the safety and immunogenicity of the rVSV-Ebola vaccine in Europe and Africa. Sci. Transl. Med. 9, eaaj1701 (2017).
Conti, P. et al. Activation of human natural killer cells by lipopolysaccharide and generation of interleukin-1 alpha, beta, tumour necrosis factor and interleukin-6. Effect of IL-1 receptor antagonist. Immunology 73, 450–456 (1991).
Cifaldi, L. et al. Inhibition of natural killer cell cytotoxicity by interleukin-6: implications for the pathogenesis of macrophage activation syndrome. Arthritis Rheumatol. 67, 3037–3046 (2015).
Biron, C. A., Nguyen, K. B., Pien, G. C., Cousens, L. P. & Salazar-Mather, T. P. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 17, 189–220 (1999).
Agnandji, S. T. et al. Phase 1 trials of rVSV Ebola vaccine in Africa and Europe—preliminary report. N. Engl. J. Med. 347, 1647–1660 (2015).
Nguyen, K. B. et al. Coordinated and distinct roles for IFN-αβ, IL-12, and IL-15 regulation of NK cell responses to viral infection. J. Immunol. 169, 4279–4287 (2002).
Wu, Y., Tian, Z. & Wei, H. Developmental and functional control of natural killer cells by cytokines. Front. Immunol. 8, 930 (2017).
Didierlaurent, A. M. et al. Enhancement of adaptive immunity by the human vaccine adjuvant AS01 depends on activated dendritic cells. J. Immunol. 193, 1920–1930 (2014).
Jensen, H., Andresen, L., Nielsen, J., Christensen, J. P. & Skov, S. Vesicular stomatitis virus infection promotes immune evasion by preventing NKG2D-ligand surface expression. PLoS ONE 6, 1–8 (2011).
Strandmann, E. P., von, Shatnyeva, O. & Hansen, H. P. NKp30 and its ligands: emerging players in tumor immune evasion from natural killer cells. Ann. Transl. Med. 3, 314 (2015).
Santoro, F. Transcriptomic data deposit for Pejoski et al. npj Vaccines https://doi.org/10.5281/zenodo.3415147 (2020).
Pejoski, D. Data depository for Pejoski et al. npj Vaccines https://doi.org/10.6084/m9.figshare.11886678 (2020).
Acknowledgements
We would like to acknowledge the individual members of the VEBCON Consortium who enabled the clinical trials, and the VSV-EBOVAC and VSV-EBOPLUS research consortia for their intellectual input, sharing of reagents and data, and funding contributions. We also thank the volunteers of the trial for their participation. The VSV-EBOVAC and VSV-EBOPLUS Consortia are supported by the IMI2 Joint Undertaking (www.imi.europa.eu) within the IMI Ebola+ program launched in response to the EVD outbreak in Western Africa in 2014. The phase 1 clinical trial was funded by the Wellcome Trust Foundation through a grant awarded to the WHO.
Author information
Authors and Affiliations
Consortia
Contributions
C.-A.S. and A.H. designed the clinical trial; all authors contributed to either experimental or analysis design; C.D., D.P., P.M.M., F.S., M.H. performed the experiments and/or analysis; D.P. and C.-A.S. wrote the paper alongside editing contributions from all authors. The VEBCON consortium enabled the clinical trials, while the VSV-EBOVAC and VSV-EBOPLUS Consortia made substantial contributions to the conception and design of the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pejoski, D., de Rham, C., Martinez-Murillo, P. et al. Rapid dose-dependent Natural Killer (NK) cell modulation and cytokine responses following human rVSV-ZEBOV Ebolavirus vaccination. npj Vaccines 5, 32 (2020). https://doi.org/10.1038/s41541-020-0179-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41541-020-0179-4