Abstract
When the proliferation of residual hepatocytes is prohibited, biliary epithelial cells (BECs) transdifferentiate into nascent hepatocytes to accomplish liver regeneration. Despite significant interest in transdifferentiation, little is known about the maintenance of nascent hepatocytes in post-injured environments. Here, we perform an N-ethyl-N-nitrosourea (ENU) forward genetic screen and identify a mutant containing a nonsense mutation in the gene nibrin (nbn), which encodes a component of the Mre11-Rad50-Nbn (MRN) complex that activates DNA damage response (DDR). The regenerated hepatocytes cannot be maintained and exhibit apoptosis in the mutant. Mechanistically, the nbn mutation results in the abrogation of ATR-Chk1 signaling and accumulations of DNA damage in nascent hepatocytes, which eventually induces p53-mediated apoptosis. Furthermore, loss of rad50 or mre11a shows similar phenotypes. This study reveals that the activation of DDR by the MRN complex is essential for the survival of BEC-derived hepatocytes, addressing how to maintain nascent hepatocytes in the post-injured environments.
Similar content being viewed by others
Introduction
The liver is a vital organ with a unique regenerative capacity. Upon varying degrees of injury, the regenerating liver parenchymal cells are derived from pre-existing hepatocytes or biliary epithelial cells (BECs)1,2,3,4,5. Many studies have reported that ductular reactions occur in certain pathological conditions such as viral hepatitis, alcoholic hepatitis, nonalcoholic steatohepatitis, and liver cirrhosis6,7. Recent findings further show that BECs extensively contribute to regenerated hepatocytes upon severe liver injury in zebrafish8,9 and mice10,11,12,13 when hepatocyte-mediated regeneration is compromised. BECs dedifferentiate into bi-potential progenitor cells (BPPCs) and subsequently redifferentiate and proliferate into hepatocytes and BECs in zebrafish8,9. Considering the crucial roles of hepatocytes in physiological functions, it is necessary to explore the mechanisms underlying the formation and survival of regenerated hepatocytes for the clinical therapeutics of liver diseases.
Liver regeneration is a complex and highly regulated process that ensures damaged hepatic structural and functional restoration. When hepatocytes are severely injured, or the proliferative capacity of residual hepatocytes is inhibited in zebrafish or mouse models8,9,10,11,12,13, nascent hepatocytes mainly originate from BECs. Given the conservation between both species, understanding the mechanisms underlying the generation and maintenance of regenerated hepatocytes will be useful for elucidating the regulatory mechanisms underlying liver regeneration in end-stage liver diseases. Recently, our group addressed that the mammalian target of rapamycin complex 1 signaling and DNA methyltransferase 1 are required for the BEC-to-BPPC dedifferentiation after extreme liver damage14,15. Other studies have suggested farnesoid X receptor-extracellular signal-related kinase 1 axis, Wnt, and bone morphogenetic protein govern the BPPC-to-hepatocyte redifferentiation16,17,18, and Notch regulates the BPPC-to-BEC redifferentiation19. Although many factors involved in cell transdifferentiation have been identified, the molecular mechanisms underlying the maintenance of regenerated hepatocytes remain largely unknown.
The MRN complex, composed of Mre11, Rad50, and Nbn, is involved in the maintenance of genomic stability, DNA repair, and cell cycle regulation. Besides, the MRN complex is an extensively documented sensor of DNA damage response (DDR) that detects DNA double-strand breaks (DSBs) and replication stress to fully activate both ataxia telangiectasia mutated (ATM) and RAD3-related (ATR)-dependent signalings20,21. ATM and ATR are the key mediators of the DDR. Upon recruitment to the sites of DNA damage, ATM and ATR phosphorylate downstream substrates Chk2 and Chk1, respectively, to induce cell cycle arrest and initiate DNA repair22,23,24,25. Subsequently, p53 is activated to allow repair of DNA damage or undergo apoptosis if DNA damage is irreparable. Although the MRN complex plays an important role in DDR, its function in liver regeneration, particularly in extreme hepatocyte injury, has not been addressed.
For the first time, our group undertook an N-ethyl-N-nitrosourea (ENU)-mutagenesis-based forward genetic screen of liver regeneration in zebrafish to explore the mechanisms underlying liver regeneration after extreme hepatocyte loss. We identified a mutation in nibrin (nbn) by positional cloning and reverse genetics approaches, which shows severe defects during the later stage of liver regeneration, but does not affect the transdifferentiation process. Further, we used rad50 mutant and chemical inhibition of ATM, ATR, or Chk1 to reveal MRN-ATR-Chk1 axis is indispensable for the survival of regenerated hepatocytes. Besides, cell apoptosis induced by impaired MRN complex is p53-dependent during liver regeneration in vivo. In summary, this study suggests the essential role of MRN complex in maintaining the survival of hepatocytes during BECs-driven liver regeneration.
Results
The zebrafish lvv cq137 mutant displays defective maintenance of nascent hepatocytes in liver regeneration after extreme liver injury
To explore novel genes and regulatory mechanisms in liver regeneration, we carried out an N-ethyl-N-nitrosourea (ENU) forward genetics screen under the Tg(lfabp:DenNTR)cq1 background8, in which liver fatty acid-binding protein 10a (lfabp) promoter drives the expression of Dendra2 and nitroreductase (NTR) fusion protein in hepatocytes. The Tg(lfabp:DenNTR) transgenic larvae were treated with 10 mmol/L metronidazole (Mtz) for 24 hours from 5 days post-fertilization (dpf) (Fig. 1a). Then, hepatocyte regeneration could be prominently observed at regeneration 24 hours (R24h) after withdrawal of Mtz. The regenerating liver restored 50% of its normal size at R48h and fully regenerated within the next two days8,9. After the screen, we identified a recessive mutant named liver victor (lvv)cq137, which exhibited relatively normal liver size at R24h but dramatically reduced at R48h in contrast to the siblings (Fig. 1b, c). Overall, no obvious developmental defects were observed in the lvv mutant (Fig. 1d). To confirm the defect of liver regeneration in lvv mutant, we further examined the expressions of hepatic markers, such as betaine-homocysteine methyltransferase (bhmt), ceruloplasmin (cp), and vitamin D binding protein (gc) by whole-mount in situ hybridization (WISH). The expression intensity and area of bhmt, cp, and gc were comparable between mutants and siblings at R24h (Fig. 1e). In contrast, the expression area of these markers substantially decreased in the mutants at R48h (Fig. 1e). These results suggest the maintenance of nascent hepatocytes is defective during liver regeneration in lvv mutant.
a Schematic illustration of Mtz treatment and analysis at BT (5 dpf), R0h (6 dpf), R24h (7 dpf), and R48h (8 dpf). b Confocal projection images showing the regenerating livers of sibling and lvv mutant from BT to R48h under the Tg(lfabp:DenNTR) background. c Quantification of the area of liver sizes, n = 7 larvae per group. d Bright-field and epifluorescence images showing the morphological appearance and regenerating livers (arrows) of sibling and lvv larvae at R48h, respectively. e WISH images showing the expressions of bhmt, cp, and gc in regenerating livers (arrows) at R24h and R48h in sibling and lvv larvae. Asterisks indicate statistical significance: ****P < 0.0001 using t-tests analysis when compared to control. Numbers indicate the proportion of larvae exhibiting the phenotype shown. Scale bars, 100 μm b, e, and 300 μm d; error bars, ±SEM. BT before Mtz treatment, R regeneration time after the withdrawal of Mtz, WISH whole-mount in situ hybridization, n.s. no significance.
To exclude the developmental defect of BECs and bile secretion in lvv mutant, we performed a feeding assay with the fluorescent BODIPY FL C5, a fatty acid analogue that helps to visualize the bile secretion of hepatocytes and the bile conduction in bile ducts26. Normal accumulation of BODIPY FL C5 in the bile ducts and gallbladder at 5 dpf indicated that hepatocytes and BECs are functional in lvv mutant (Supplementary Fig. 1a). We also examined the morphology of bile ducts using the Tg(Tp1:Tomato) line15,16, and found the biliary duct system was well developed in lvv mutant (Supplementary Fig. 1b). To confirm whether BEC transdifferentiation was disturbed in lvv mutant, the Cre/loxP-mediated lineage tracing was performed using the Tg(krt18:CreER) transgenic line14,15. Validated by the Tg(krt18:CreER; βactin:loxP-DsRed-loxP-GFP) (Supplementary Fig. 2a, b), almost all the nascent hepatocytes derived from the transdifferentiation of BECs in the Tg(krt18:CreER; lfabp:loxP-STOP-loxP-DsRed; lfabp:DenNTR) line in sibling and lvv mutant (Supplementary Fig. 2c, d). These data suggest that the origins of nascent hepatocytes are unaffected in lvv mutant.
As the mutant exhibited normal liver sizes from 5 to 8 days post-fertilization (dpf) (Supplementary Fig. 3a), we further checked the development of digestive organs using ceruloplasmin (cp), prospero homeobox 1a (prox1a), fatty acid binding protein 2 (fabp2), trypsin, and insulin probes. WISH results showed that the liver, intestine, and pancreas develop normally in lvv mutant (Supplementary Fig. 3b, c). These results indicate that the lvv mutant explicitly affects liver regeneration without obvious developmental defects.
Liver regeneration defect in lvv mutant is caused by the mutation of nbn
To identify the target gene of lvv, we used the positional cloning approach to map the mutation site. The mutation was localized within seven candidate genes flanked by two closely linked simple sequence length polymorphism (SSLP) markers, L16m1 and L16m2, on chromosome 16 (Fig. 2a). After sequencing with cDNA and genomic DNA in lvv and sibling, we identified a candidate mutation in the nbn gene within this linked region. The T to A nonsense mutation was identified (Leu to STOP coding transition) and predicted to truncate Nbn prior to the putative nuclear localization signal (NLS) and DNA repair domain; thus, the mutation is predicted to be a loss-of-function allele (Fig. 2b, c). Then we analyzed the subcellular localization of Nbncq137. A Myc-tag was fused to the N-terminus of wild-type Nbn (NbnWT) protein and mutated Nbn (Nbncq137) protein for detection, respectively (Fig. 2d). Confocal images showed that Myc-tagged NbnWT was predominantly localized in the nucleus of regenerated hepatocytes (Fig. 2e). However, Myc-tagged Nbncq137 was mistakenly localized in the cytoplasm (Fig. 2e), which is consistent with the lack of putative NLS in Nbncq137. These results suggest that the Nbncq137 cannot form the nuclear foci and loses its nuclear function of DNA repair. It has been reported that Nbn is mainly expressed in regions of high proliferative activity or organs with physiologic DSBs27. We examined the spatiotemporal pattern of nbn expression. WISH assay showed the expression of nbn mRNA was gradually upregulated from R8h to R48h in the liver region during liver regeneration (Fig. 2f), which indicates nbn plays a role in the later stage of liver regeneration.
a Genetic map of the candidate region on chromosome 16. Numbers above SSLP markers indicate recombination events. b Sequencing results displaying the mutation site in nbn. c Schematic diagram of the structure of Nbn protein. FHA, BRCT, and NLS denote forkhead-associated domain, BRCA1 C-terminal domain, and nuclear localization signal, respectively. d Schematic illustration of Mtz treatment and analysis at R48h. e Single optical section images showing Myc (red), Dendra2 (green), and DAPI (blue) staining in regenerating livers at R48h under the Tg(lfabp:DenNTR) background. Note that Myc-tagged NbnWT is mainly localized in the nucleus, while Myc-tagged Nbncq137 is localized in the cytoplasm. f WISH images showing nbn expression in the liver region (arrows) from BT to R48h. Numbers indicate the proportion of larvae exhibiting the expression shown. Scale bars, 100 μm. WT wild-type, DAPI 4′, 6-diamidino-2-phenylindole, BT before Mtz treatment, R regeneration time after the withdrawal of Mtz, WISH whole-mount in situ hybridization.
To confirm the disruption of nbn was causative for the phenotype of lvv mutant, we generated a new nbn mutant line nbncq139 using the CRISPR/Cas9 system. Due to the ENU nonsense mutation causing premature termination of the Nbn protein, we designed two specific gRNAs based on the sequences of nbn in exon 11 and exon 15 for large fragment deletion to mimic the ENU mutation (Supplementary Fig. 4a). Wild-type alleles were too long to be amplified under conventional PCR conditions, while the knockout produces a 500 bp fragment that can be separated by gel electrophoresis (Supplementary Fig. 4b). The nbncq139 mutant phenocopied the liver regeneration defects of lvv; a relatively normal liver size was formed at R24h, and the liver size decreased dramatically at R48h (Supplementary Fig. 4c, d). To further confirm whether the disrupted nbn was responsible for the defective liver regeneration, Tg(hsp70l:nbn-p2A-mCherry)cq141 was generated for overexpression of Nbn in the lvv mutant after heat-shock induction (Supplementary Fig. 4e). Liver regeneration defect in lvv mutant could be rescued by Nbn overexpression, which was identified by the red fluorescence of mCherry (Supplementary Fig. 4e, f). These results validate that the phenotype of impaired liver regeneration in lvv mutant is caused by nbn mutation.
Nbn maintains liver regeneration depending on the function of MRN complex
Coincidentally, we identified another ENU-induced mutant cq138 that showed dramatic liver size reduction at R48h in contrast to the sibling, while the liver size had no significant difference between siblings and cq138 mutant at R24h (Fig. 3a). In general appearance, no obvious developmental defects were observed in the cq138 mutant (Fig. 3b). In addition, the development of digestive organs such as the liver, intestine, and pancreas in cq138 mutant were normal (Supplementary Fig. 5). These results indicate that the cq138 mutant does not cause obvious developmental defects but specifically affects the process of the later stage of liver regeneration, which phenocopies the nbn mutant.
a Confocal projection images showing the regenerating livers of sibling and cq138 mutant from BT to R48h under the Tg(lfabp:DenNTR) background. b Bright-field and epifluorescence images showing the morphological appearance and regenerating livers (arrows) of sibling and cq138 larvae at R48h, respectively. c Sequencing result displaying the mutation site in rad50. d Schematic illustration of Mtz treatment and analysis at R48h. Single optical section images showing Myc (red), Dendra2 (green), and DAPI (blue) staining in regenerating livers of wild-type and cq138 mutant at R48h under the Tg(lfabp:DenNTR) background. Note that Myc-tagged NbnWT is mislocalized to the cytoplasm in rad50-/- larvae. e Confocal projection images showing the regenerating livers of wild-type and nbn-/- rad50-/- double mutant from BT to R48h under the Tg(lfabp:DenNTR) background. f Schematic illustration of HU and mirin treatment and analysis at R48h. Confocal projection images showing the regenerating livers of larvae treated with or without mirin under HU exposure. Numbers indicate the proportion of larvae exhibiting the expression shown. Scale bars, 100 μm (a, d, e, and f) and 300 μm b. BT before Mtz treatment, R regeneration time after the withdrawal of Mtz, WT wild-type, DAPI 4′, 6-diamidino-2-phenylindole, HU hydroxyurea.
We identified rad50, another component of the MRN complex, as the mutation gene of cq138 by positional cloning and sequencing. A point mutation from T to A replacement in the exon 25 of the rad50 resulted in an Ile to Asn conversion (Fig. 3c). To further determine the identity of the mutated gene in cq138, we generated another mutant of cq140 that carried a 6 bp deletion and 7 bp insertion by targeting the exon 15 of rad50 using CRISPR/Cas9 technology (Supplementary Fig. 6a, b). Rad50cq140 could well recapitulate the phenotype of ENU-induced rad50cq138 (Supplementary Fig. 6c, d).
Rad50, together with Nbn and Mre11, forms an evolutionary conserved MRN complex in vertebrates28,29,30. To examine whether nbn and rad50 act in a complex manner during liver regeneration, we analyzed the subcellular localization of Myc-tagged NbnWT in rad50 mutant. Confocal imaging showed that Myc-NbnWT was mislocalized in the cytoplasm in rad50 mutant (Fig. 3d). Since Myc-Nbncq137 was cytoplasmic in wild-type (Fig. 2e) and Myc-NbnWT could not form nuclear foci in rad50 mutant (Fig. 3d), we inferred that recruitment of the MRN complex to the DNA damage site was disturbed in both nbn and rad50 mutant larvae. Next, we intercrossed nbn and rad50 heterozygotes to generate a double mutant. The nbn-/- rad50-/- double mutant exhibited identical liver regeneration defects as nbn or rad50 mutant (Fig. 3e), indicating that the effects of nbn and rad50 were not additive. Furthermore, we observed that the addition of mirin, a characterized inhibitor of Mre11 nuclease activity, to hydroxyurea (HU) exposure, which could trigger replication fork stalling and induce DNA damage31,32, showing a phenotype similar to nbn mutation and rad50 mutation (Fig. 3f). All three components of the MRN complex were indispensable for liver regeneration. These results suggest that nbn, rad50, and mre11a work as a complex in liver regeneration, and the integrity of the MRN complex is required to maintain nascent hepatocytes in liver regeneration.
Loss of nbn induces cell apoptosis of nascent hepatocytes
Liver regeneration after extreme hepatocyte loss mainly includes dedifferentiation of BECs into BPPCs and proliferation and redifferentiation of BPPCs into nascent hepatocytes and BECs8,9. To investigate how nbn affected the maintenance of regenerated hepatocytes, we first detected the expressions of the hepatoblast and BPPC markers forkhead box A3 (foxa3), hematopoietically expressed homeobox (hhex), and SRY-box transcription factor 9b (sox9b), all of which were induced in BPPCs during liver regeneration14,16. The transcriptional activation of foxa3, hhex, and sox9b in nbn mutant was comparable to wild-type at R0h (Supplementary Fig. 7a), indicating that the process of dedifferentiation is undisturbed. Given that the regenerating liver in nbn mutant was relatively normal at R24h but significantly diminished at R48h (Fig. 1b, c), we doubted that the reduced liver regeneration is possibly attributable to decreased cell proliferation or increased cell death. We examined the expression of proliferation markers during liver regeneration. We performed a 5-ethynyl-20-deoxyuridine (EdU) incorporation assay. EdU and Dendra2 double immunostaining results indicated no significant difference in the percentage of EdU and Dendra2 double-positive cells between wild-type and nbn mutant larvae at R24h and R48h (Fig. 4a, b). Proliferating cell nuclear antigen (PCNA) immunostaining also displayed that the proportion of PCNA among Dendra2 double-positive cells showed no difference between wild-type and nbn mutant larvae (Supplementary Fig. 7b, c). We then performed a TdT-mediated dUTP nick-end labelling (TUNEL) assay to detect cell apoptosis. TUNEL assay showed a significant increase of apoptotic Dendra2 positive cells in nbn mutant, compared with wild-type at R24h and R48h (Fig. 4c, d). Above all, these results suggest that loss of nbn stimulates cell apoptosis, which accounts for the abrogation of regenerated hepatocytes.
a Confocal z-stack projection images showing the label of EdU (red), Dendra2 (green), and DAPI (blue) in regenerating livers at R24h and R48h in wild-type and nbn mutant. b Quantification of the percentage of EdU+ cells among Dendra2+ cells, n = 7 larvae per group. c Confocal z-stack projection images showing the label of TUNEL (red), Dendra2 (green), and DAPI (blue) in regenerating livers at R24h and R48h. d Quantification of the percentage of TUNEL+ cells among Dendra2+ cells, n = 7 larvae per group. Asterisks indicate statistical significance: ****P < 0.0001 using t-tests analysis when compared to control. Scale bars, 100 μm; error bars, ±SEM. R regeneration time after the withdrawal of Mtz, WT wild-type, DAPI 4′, 6-diamidino-2-phenylindole, n.s. no significance.
Accumulative DNA damage and impaired ATR-Chk1 signaling in nascent hepatocytes in nbn mutant during liver regeneration
Nbn is one of the main components of MRN complex, which acts as a central sensor of DSBs and plays a role in preventing replication stress for faithful DNA replication20,21. Complete deletion of any component of the MRN complex is incompatible with life in mice, presumably due to replication-associated DSBs33,34,35,36. To evaluate the level of endogenous DNA damage in nbn mutant, we measured the typical DNA damage marker, phosphorylated H2AX (γH2AX). Although γH2AX was rarely detected in siblings, γH2AX positive cells were significantly increased and accumulated in the nbn mutant liver at R24h and R48h (Fig. 5a). TUNEL assay combined with γH2AX staining further revealed that cells with accumulated DNA damage undergo apoptosis (Supplementary Fig. 8). The increased number of γH2AX positive cells in nbn mutant larvae suggested that disruption of nbn leads to accumulation of endogenous DNA damage in regenerating liver during rapid proliferation of hepatocytes.
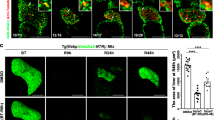
a Single optical section images showing γH2AX (red), Dendra2 (green), and DAPI (blue) expressions in regenerating livers at R24h and R48h in wild-type and nbn mutant. Quantification of the number of γH2AX+ cells per liver, n = 7 larvae per group. b Schematic illustration of Mtz treatment and inhibitor treatment under HU exposure. c Western blot images of p-Chk1 in wild-type, HU-treated wild-type and HU-treated nbn mutant larvae. Note that nbn mutation abrogates the Chk1 phosphorylation upon HU treatment. d Western blot images of p-Chk2 in wild-type, HU-treated wild-type, and HU-treated nbn mutant larvae. e Confocal projection images showing the regenerating livers of Chk1 inhibitor treatment under the Tg(lfabp:DenNTR) background. f Confocal projection images showing the regenerating livers of ATR inhibitor and ATM inhibitor treatment under the Tg(lfabp:DenNTR) background. Note that ATR inhibitor but not ATM inhibitor treatment shows reduced liver regeneration in wild-type upon HU treatment. Asterisks indicate statistical significance: **P < 0.01; ***P < 0.001 using t-tests analysis when compared to control. Numbers indicate the proportion of larvae exhibiting the expression shown. Scale bars, 100 μm; error bars, ±SEM. WT wild-type, R regeneration time after the withdrawal of Mtz, DAPI 4′, 6-diamidino-2-phenylindole, HU hydroxyurea, Chk1i Chk1 inhibitor CHIR-124, ATRi ATR inhibitor VE-821, ATMi ATM inhibitor KU55933.
All living cells are constantly challenged by damaging agents of endogenous and external stresses. Numerous highly conserved DDR pathways exist to activate DNA damage repair and cell cycle checkpoints37,38. ATM and ATR are central to the DDR; when activated, both trigger a series of downstream signaling events, including Chk2 and Chk1, respectively39,40. In the case of DNA damage in rapidly proliferating cells, we evaluated the activation of DNA damage checkpoints in nbn mutant by western blotting. We checked phosphorylation levels of Chk1 and Chk2 in response to HU exposure (Fig. 5b), which could trigger replication fork stalling and induce DNA damage31,32. Notably, the phosphorylation level of Chk1 at Ser345 (p-Chk1) in wild-type was increased at R48h after 100 mM HU treatment. However, the activation of p-Chk1 in nbn mutant was largely compromised (Fig. 5c and Supplementary Fig. 9a), indicating that disruption of Nbn inhibits the Chk1 signaling during liver regeneration. In contrast, the phosphorylation level of Chk2 at Thr68 (p-Chk2) was comparable in nbn mutant and wild-type under HU treatment (Fig. 5d and Supplementary Fig. 9b), indicating that Chk2 signaling remains functional in nbn mutant. To verify whether Chk1 plays a role in liver regeneration, we treated the regenerating liver with the Chk1 inhibitor CHIR-124 after HU exposure. The liver size of CHIR-124 treatment was significantly decreased after HU exposure, representing the phenotype of nbn mutant (Fig. 5e). Collectively, these data indicate that Nbn deficiency impairs Chk1 signaling in response to the endogenous DNA damage of the regenerating liver.
To better understand the role of DDR in liver regeneration, we examined the role of ATR and ATM signaling with two specific kinase inhibitors, VE-821 and KU55933. Tg(lfabp:DenNTR) larvae treated with 5 µM ATR inhibitor (VE-821) from R8h to R48h showed a much smaller liver compared with the control group under HU exposure, while the liver was relatively normal after inhibition of ATM (Fig. 5f). In addition, the treatment with these inhibitors did not lead to apoptosis of hepatocytes during development (Supplementary Fig. 10). Since ATR inhibition and Chk1 inhibition mimicked the phenotype of nbn mutant, we inferred that a failure of ATR-Chk1 signal activation mainly caused the liver regeneration defects. Above all, these data suggest that the blockage of the Nbn-ATR-Chk1 axis is associated with the defective regenerated hepatocytes in nbn mutant.
P53 mediates hepatocyte apoptosis in nbn mutant during liver regeneration
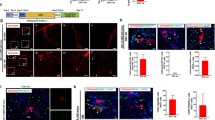
As p53 regulates cell cycle progression and apoptosis in response to DNA damage41,42,43, we examined the expressions of several p53 target genes to check whether the increased cell apoptosis in nbn mutant was triggered by aberrant activation of p53. The WISH results showed that the transcription activation of p53, p21, and mdm2 were significantly upregulated in nbn mutant at R24h and R48h (Fig. 6a). The upregulation of p21 should result in decreased cell proliferation as previously reported11, and then we found the percentage of EdU and Dendra2 double-positive cells was reduced in nbn mutant larvae compared with wild-type at R72h and R96h (Supplementary Fig. 11). It implied the reduced proliferation was cumulative in time and appeared in nbn mutant during the later stage of liver regeneration. Additionally, the origin of nascent hepatocytes did not change over time and remained from the transdifferentiation of BECs (Supplementary Fig. 12). To further confirm that the upregulation of p53 signaling was responsible for apoptosis in nbn mutant, we introduced tp53M214K mutant (p53-/-) into the nbn mutant, which abrogated normal p53 function in apoptosis44. Loss of p53 did not affect liver regeneration as previously described (Supplementary Fig. 13)15. The size of regenerating liver in nbn mutant was partially rescued in p53 heterozygous, and the rescue effect was more obvious in p53 homozygous mutant (Fig. 6b, c). Further study showed that cell apoptosis was significantly decreased at R48h in nbn-/- p53-/- double mutant compared with nbn mutant embryos (Fig. 6d, e). Besides, the expressions of hepatocyte markers were well restored in nbn-/- p53-/- double mutant compared with nbn mutant by the WISH results of bhmt, cp and gc (Fig. 6f). The expression of p21 was downregulated in nbn-/- p53-/- double mutant compared to nbn mutant, suggesting that p21 acts in a p53-dependent manner (Supplementary Fig. 14). In summary, the cell apoptosis of regenerating hepatocytes in nbn mutant is p53-dependent.
a WISH result showing the expressions of p53, mdm2, and p21 in wild-type and nbn mutant livers at R24h and R48h (arrows). b, c Three kinds of standards of liver regeneration b and quantitative analysis of regeneration phenotype c. Note that inactivation of p53 can partially rescue the liver regeneration in nbn mutant. The numbers of embryos are shown above the columns. d Confocal z-stack projection images showing the label of TUNEL (red), Dendra2 (green), and DAPI (blue) in wild-type, nbn-/-, and nbn-/- p53-/- double mutant regenerating livers at R48h. Note that cell apoptosis is decreased in nbn mutant by introducing p53-/-. e Quantification of the area of liver sizes and the number of TUNEL+ cells per liver, n = 6 larvae per group. f WISH images showing the expressions of bhmt, cp, and gc in wild-type, nbn-/-, and nbn-/- p53-/- double mutant regenerating livers at R48h (arrows). Asterisks indicate statistical significance: ****P < 0.0001 using t-tests analysis when compared to control. Numbers indicate the proportion of larvae exhibiting the expression shown. Scale bars, 100 μm; error bars, ±SEM. R regeneration time after the withdrawal of Mtz, WT wild-type, WISH whole-mount in situ hybridization, DAPI 4′, 6-diamidino-2-phenylindole, n.s. no significance.
The MRN complex is required for the survival of regenerating hepatocytes in zebrafish fibrotic liver model and partial hepatectomy in mice
To study the contribution of MRN complex in other forms of liver regeneration, we applied extreme liver injury in ethanol (EtOH)-induced fibrotic liver zebrafish model17. Larvae were exposed to 1.5% EtOH for inducing liver fibrosis (Supplementary Fig. 15a). The nbn mutant exhibited liver regeneration defects and increased cell apoptosis in fibrotic liver injury model (Supplementary Fig. 15b and c). These results suggest the MRN complex plays a role in the liver regeneration of the fibrotic liver injury model.
To further extend the significance of our findings, we further checked the roles of MRN complex in liver regeneration using a partial hepatectomy (PHx) model in mice (Supplementary Fig. 16a). We found a reduction in the liver-to-body weight ratio (Supplementary Fig. 16b) and a significant increase of apoptotic hepatocytes (Supplementary Fig. 16c) with the Chk1 inhibitor CHIR-124 after HU exposure. Besides, CHIR-124 treatment increased the activity of serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) while decreasing serum albumin concentration compared to the control group (Supplementary Fig. 16d). Taken together, these results indicate the MRN complex has a conserved function that regulates the maintenance of nascent hepatocytes in mouse PHx model as in zebrafish.
Discussion
During liver regeneration, BPPCs undergo rapid cell proliferation and inevitably face an increased risk of replication-associated spontaneous DNA damage and stalled replication fork, referred to as DNA replication stress, which is required to be efficiently resolved by robust DNA damage response (DDR). Cell cycle arrest is required to provide time for DNA repair and replication fork restart; If fork restart fails, replication forks collapse into DSBs45,46,47,48. The function of DDR in liver regeneration after severe liver injury remains largely unknown. Here, using a zebrafish extreme liver injury model with an ENU-based forward genetic screen, we identified a novel mutant with relatively normal liver development but exhibiting severe liver regeneration defects, thus revealing previously unreported roles of DDR factor nbn. Mechanically, recruitment of Nbn can activate ATR kinase activity and then phosphorylate downstream target Chk1. Activated Chk1 promotes replication forks stabilization and restart during liver regeneration so that the regenerated hepatocytes can survive. Moreover, we described that disrupting the function of Rad50 and Mre11a mimics the phenotype of nbn mutant. Taken together, we uncovered that nbn activates the ATR-Chk1 axis through MRN complex, which is indispensable for the survival of regenerated hepatocytes (Fig. 7).
During the highly proliferative phase of liver regeneration, the hepatocytes undergo endogenous DNA damage caused by replication stress. In normal livers, the MRN complex is recruited to DNA damage and activates ATM and ATR, subsequently triggering an intricate signal transduction network called DNA damage response. Activated cell cycle checkpoints arrest cell cycle progression and allow DNA repair so regenerated hepatocytes can survive and proliferate. In nbn mutant, the truncated Nbn mislocalizes in the cytoplasm, and the MRN complex cannot be recruited to the DNA damage site. The ATR-Chk1 signaling is prevented from activation. The stalled replication fork may collapse and trigger p53-dependent apoptosis, ultimately leading to defective liver regeneration.
Traditionally, the MRE11/RAD50/NBN deficiency causes chromosome instability syndromes in humans28,49,50. The patient may display growth retardation, immunodeficiency, and predisposition to cancer. Cells from patients exhibited increased sensitivity to ionizing radiation and increased levels of spontaneous chromosomal fragility33,49,51. In mice, the disruption of either component of the MRN complex causes early embryonic lethality, which makes it difficult to uncover the disease-associated pathology and related molecular mechanism in the mouse model34,36,52. With in-depth analysis during zebrafish development, we found the larvae of nbn or rad50 mutants exhibited relatively normal morphological appearance. To test whether the homozygous mutant animals can survive to adulthood, we genotyped the progenies generated from heterozygous parents at 1 month post-fertilization (mpf) and 4 mpf. Homozygous mutants could be genotyped at 1 mpf but completely failed to survive to 4 mpf. Intriguingly, we also found that the nbn and rad50 mutant zebrafish showed significant growth retardation relative to siblings, as zebrafish mre11a mutant showed growth defects and failed to survive to adult stages53. That is consistent with human Nijmegen Breakage Syndrome. Thus, our findings suggest that Nbn or Rad50 deficiency zebrafish may afford an opportunity to understand the mechanism of chromosome instability syndromes in humans.
MRN complex is recognized as one of the first sensors that respond to DSB or replication stress. Stimulated by DSB or replication fork arrest, MRN complex recruit and activate ATM and ATR kinases. Then, Chk2 and Chk1 are phosphorylated to coordinate DNA repair and cell cycle progression22. Studies on nbn mutants showed that DNA damage accumulation and apoptosis increase in the regenerated hepatocytes, which is related to the failed MRN/ATR/Chk1 axis activation. During the rapid proliferative phase of liver regeneration, the importance of DDR for nascent hepatocyte survival is further emphasized by hydroxyurea treatment.
In our study, treatment with selective inhibitors of MRN/ATR/Chk1 axis did not induce hepatocyte apoptosis during zebrafish development, indicating that these small molecules were non-toxic at the doses we used. These small molecules have been verified to be highly selective previously54 and were used at concentrations of 10 μM of ATMi and ATRi in cancer cells55. However, studies of ATM/ATR inhibitors in zebrafish remain limited. Therefore, we explored the dosages of ATMi and ATRi in zebrafish and finally determined the appropriate doses for treating the larvae with 2 μM and 5 μM, respectively. However, due to the general disadvantage of small molecules of lacking exclusive selectivity, additional genetic validation or further mechanistic studies are needed.
Furthermore, using a zebrafish fibrotic liver model, we found the nbn mutants exhibited liver regeneration defects and increased hepatocyte apoptosis. Similarly, using a mouse partial hepatectomy model, we also found a defective hepatic regeneration and a significant increase of apoptotic hepatocytes with inhibition of the function of MRN complex. These results indicate that the role of MRN complex is also involved in maintaining the survival of nascent hepatocytes in other forms of liver injuries, indicating the critical role of MRN complex in a broad manner of liver regeneration.
Upon activation of the DDR, p53 is activated to induce cell cycle arrest, DNA repair, or apoptosis41,42,43. How p53 chooses the outcomes between the competing cell survival and cell death upon the extent and type of DNA damage and cell type has been a great fascination but is not yet entirely elucidated. Here, we describe that the p53-p21 axis was hyperactivated in nbn mutants. Loss of p53 could partially rescue the deficiency of liver regeneration in nbn homozygous mutants. These results suggest that p53 loss of function benefits the survival of regenerated hepatocytes in nbn deficiency zebrafish. Understanding detailed molecular mechanisms underlying the regulation of MRN-mediated p53 activation awaits further investigation.
Collectively, this study revealed a novel and vital role of MRN-ATR-Chk1 signaling in regenerated hepatocytes after extreme liver injury. Loss of nbn did not affect the early phase of BEC-mediated transdifferentiation but specially regulated the survival of nascent hepatocytes through the ATR-Chk1 axis. rad50 mutants validated the phenotype of nbn mutant is a direct consequence of MRN complex dysfunction. Given the conservation roles of BECs upon severe liver injury from zebrafish to mammals, the MRN complex may be a potential target for therapeutics after severe liver injury.
Methods
The agency or committee that granted approval
All animals are housed in temperature- and light-controlled facilities and are maintained in accordance with the Guide for Care and Use of Laboratory Animal and the Animal Welfare Act. Experiments were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) at Southwest University.
Zebrafish strains
Zebrafish strains were raised and maintained under standard laboratory conditions according to Institutional Animal Care and Use Committee protocols. All experimental protocols were approved by Southwest University (Chongqing, China). The transgenic or mutant zebrafish lines Tg(lfabp:Dendra2-NTR)cq1 abbreviated as Tg(lfabp:DenNTR)cq1, Tg(Tp1:Tomato), Tg(krt18:CreER)cq74, Tg(βactin:loxP-DsRed-loxP-GFP)s928, Tg(lfabp:loxP-STOP-loxP-DsRed)cq4, Tg(hsp70l:nbn-p2A-mCherry)cq141, Tg(hsp70l:Myc-nbnWT)cq142, Tg(hsp70l:Myc-nbncq137)cq143,nbncq137 (lvv) mutant, nbncq139 mutant, rad50cq138 mutant, rad50cq140 mutant, tp53M214K mutant, and the polymorphic line SJD were used in this study. A complete list of zebrafish strains is provided in Table S1. All the strains are used as stable, germline transgenic lines in this study. Embryos were treated with 0.003% 1-phenyl-2-thiourea (PTU, Sigma-Aldrich) to inhibit pigment formation. Larvae were anesthetized in 0.16 g/L tricaine (Sigma-Aldrich) prior to experiments.
ENU mutagenesis and positional cloning of the lvv mutant
ENU mutagenesis was carried out as previously described56 for mutations affecting liver regeneration. Briefly, the adult male zebrafish of Tg(lfabp:DenNTR)cq1 were treated with 3.5 mM ENU for 1 h at weekly intervals and repeated for six times. Two weeks after the ENU treatment, those male zebrafish were outcrossed to AB females to generate F1 families. Then F1 fish were mated to Tg(lfabp:DenNTR)cq1 to generate F2 families. F2 sibling were intercrossed to generate F3 embryos for screening. The morphology of regenerating livers was analyzed at R48h after liver ablation with Mtz treatment in F3 embryos.
Heterozygous lvv fish were crossed with the polymorphic line SJD to generate the mapping population. Positional cloning was carried out as previously described56,57,58. The mutation was mapped to chromosome 16 by bulk segregation analysis with simple sequence length polymorphism (SSLP) markers Z6021 and Z3633. Through analysis of 935 mutants, the mutation was finally flanked by two designed SSLP markers, L16m1 (7 recombinants) and L16m2 (1 recombinant). The location of the lvv mutation was narrowed to a 0.5 million base pairs (Mb) genomic region. The coding sequences of candidate genes were sequenced by sibling and lvv mutant embryos, and the putative mutation was confirmed by genomic DNA sequencing. A complete list of the primers is provided in Table S2.
Generation of the nbn cq139 and rad50 cq140 mutant lines
The nbncq139 and rad50cq140 mutant lines were generated by CRISPR/Cas9 technology, as previously described59. In brief, gRNA (50 pg) and Cas9 mRNA (300 pg) were co-injected into 1-cell stage wild-type embryos, and the lysate of about 10 embryos at 48 hpf was used as templates for PCR. The PCR products were sequenced to examine potential indels that occurred in nbn and rad50 gRNA target region, and embryos with effective genome editing were raised into adults (F0). Then, F0 fish were screened to identify the founder whose progeny carried the indels, and offspring of identified F0 was raised up to get the candidate progeny (F1). Last, individual F1 adults were screened by PCR using the tail fin genomic DNA, and the genotypes of mutants were determined by DNA sequencing. The target sequence of nbn and rad50 and PCR primers are depicted in Table S2.
Generation of transgenic lines
Zebrafish nbn full-length coding sequence was cloned from 48 hpf cDNA. The primers for full-length cloning and site-directed mutagenesis are depicted in Table S2. Tg(hsp70l:nbn-p2A-mCherry; cryaa:Venus)cq141, Tg(hsp70l:Myc-nbnWT; cryaa:Venus)cq142, and Tg(hsp70l:Myc-nbncq137; cryaa:Venus)cq143 lines were produced using the pBluescript vector. Constructs flanked by the I-SceI (NEB) restriction sites were co-injected with I-SceI into zebrafish embryos of the Tg(lfabp:DenNTR)cq1 background at the 1-cell stage for transgenesis. Larvae were screened for expression of the yellow fluorescent protein (Venus) in the eyes. Tg(hsp70l:nbn-p2A-mCherry; cryaa:Venus)cq141 expressed mCherry in the whole body upon heat shock. For Tg(hsp70l:Myc-nbnWT; cryaa:Venus)cq142 and Tg(hsp70l:Myc-nbncq137; cryaa:Venus)cq143, Myc-tagged NbnWT or Nbn137 was verified by antibody staining. These zebrafish were raised and crossed to identify founder fish with germline integration, from which stable transgenic lines were established.
Whole-mount in situ hybridization and antibody staining
Whole-mount in situ hybridization were performed as previously described15,60 using the bhmt, cp, gc, prox1a, fabp2, trypsin, insulin, foxa3, hhex, sox9b, nbn, rad50, p53, mdm2, and p21 probes. A complete list of the primers is provided in Table S2. cDNA from the whole embryos at 3 dpf, R0h and R48h were used as a template for PCR to amplify the genes. Digoxygenin (Dig) labeled antisense probes were synthesized from PCR products using the T7 or SP6 RNA polymerase (Roche). Larvae were fixed with 4% paraformaldehyde (PFA) in PBS at 4 °C overnight, followed by dehydration and storage in 100% methanol at -20 °C for at least 24 h. Then the larvae were continuously rehydrated from 75%, 50%, 25%, to 0% methanol in PBT (0.1% Tween in PBS). After five times wash with PBT in 1 h, the larvae were prehybridized in HYB (50% formamide, 5 × SSC, 0.1% Tween-20, 5 mg/ml torula yeast RNA, 50 mg/ml heparin) at 67 °C for 3–5 h. Replace the HYB with Dig-labeled probes diluted in HYB and hybridize overnight at 67 °C. Remove the probes and wash successively with 100%, 75%, 50%, 25%, and 0% HYB in 2 × SSCT at 67 °C, and finally with 0.2 × SSCT. Afterward, the larvae were serially replaced with 25%, 50%, 75%, and 100% MABT (150 mM maleic acid, 100 mM NaCl, 0.1% Tween-20, pH 7.5) in 0.2 × SSCT, and blocked for 2 hours with 2% Block Reagent (Roche) in MABT. The larvae were then incubated with anti-Dig-AP antibody (Roche) at a dilution of 1:1000 with blocking buffer at 4 °C overnight. Remove the antibody and rinse the larvae with MABT for eight times. At last, the larvae were stained with NBT/BCIP solution (Roche). Images were captured using a SteREO DiscoveryV20 microscope equipped with AxioVision Rel 4.8.2 software (Carl Zeiss).
Zebrafish whole-mount antibody staining was carried out as previously described61,62,63,64. The skin of larvae was manually removed with tweezers. After five times wash with PT (1% Triton X-100 in PBS), the larvae were incubated at 4 °C overnight with the following primary antibodies: Dendra2 (1:500, ABIN361314, Antibody-online), Dendra2 (1:500, TA180094, Origene), PCNA (1:500, ab29, Abcam), γH2AX (1:500, GTX127342, Gene Tex), 2F11 (1:1000, ab71286, Abcam), GFP (1:1000, ab6658, Abcam), mCherry (1:500, ARG55723, ArigoBio), DsRed2 (1:1000, sc101526, Santa Cruz), and Myc (1:1000, ab9106, Abcam). The primary antibodies were diluted in the blocking solution (PBS + 4% BSA + 1% Triton X-100). Then, the larvae were washed eight times with PT and incubated with Alexa fluorescent-conjugated secondary antibodies diluted in the blocking solution: donkey anti-rabbit IgG Alexa fluor 488-conjugated (1:1000, A21206, Invitrogen), donkey anti-rabbit IgG Alexa fluor 647-conjugated (1:1000, A31573, Invitrogen), donkey anti-mouse IgG Alexa fluor 568-conjugated (1:1000, A10037, Invitrogen), donkey anti-mouse IgG Alexa fluor 647-conjugated (1:1000, A31571, Invitrogen), donkey anti-goat IgG Alexa fluor 568-conjugated (1:1000, A11057, Invitrogen), and donkey anti-goat IgG Alexa fluor 633-conjugated (1:1000, A21082, Invitrogen). After five washes with PT, the larvae were mounted and imaged. Images were captured using ZEN 2010 software equipped on an LSM780 or LSM880 confocal microscope (Carl Zeiss).
EdU labeling and TUNEL assay
The EdU labeling and TUNEL assay were performed as previously described8,62,65. In brief, we injected the EdU mixture (containing 0.2 mM EdU, 2% DMSO, and 4% phenol red) into the hearts of larvae and fixed the sample with 4% PFA after 1-h post-injection followed by using the Click-iT EdU Kit (Invitrogen) to label the proliferation cells. For terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay, embryos at R48h were fixed with 4% PFA. After methanol dehydration, rehydration, Proteinase K digestion, and acetone treatment, the embryos were stained with the In Situ Cell Death Detection Kit, TMR Red (Roche) at 37 °C for 2 h as described by the manufacturer.
Chemical treatment, heat shock, and BODIPY assay
For Mtz treatment, the Tg(lfabp:DenNTR) transgenic larvae at 5 dpf were incubated with 10 mM Mtz (metronidazole, Sigma) for 24 hours. For ethanol (EtOH) treatment, larvae were treated with 1.5% EtOH (Sigma Aldrich) for 48 hours, starting from 4 dpf17. For HU (hydroxyurea, Sigma) treatment, embryos were treated with 100 mM HU in egg water from R0h to R8h. Larvae were treated with 0.1 μM Chk1i (CHIR-124, Selleck), 5 μM ATRi (VE-821, Selleck), 2 μM ATMi (KU-55933, Selleck) and 20 μM Mirin (Selleck) from R8h to R48h. The chemical solutions were renewed every 24 h to maintain the pharmacological effects. And a 0.2% DMSO solution in egg water was used as a control. When chemical treatment is removed, the larvae need to be washed and recovered in the egg water.
For temporal control of CreER activities, larvae were collected and incubated with 5 μM 4-hydroxytamoxifen (4-OHT, Sigma) for 24 h from 4 to 5 dpf, followed by three washes with egg water.
The larvae of Tg(hsp70l:nbn-p2A-mCherry)cq141 were heat-shocked at 38.5 °C for 40 min at the indicated time frame and returned to 28.5 °C for further analysis.
For BODIPY feeding assay, larvae were fed with BODIPY FL C5 (Invitrogen) for 4 h at 5 dpf and then live imaging, as previously described26.
Western blotting
The western blotting was carried out as previously described61. In brief, total protein was extracted from wild-type and nbn mutant embryos (30 larvae per group) using RIPA lysis buffer containing protease inhibitor, phosphatase inhibitor and PMSF. The protein samples were separated on 10% SDS-PAGE gel and then transferred to a 0.45 μm PVDF membrane (Millipore). The membranes were blocked in 5% BSA for 2 h at the room temperature and immunoblotted with the primary antibodies as follows: β-actin (1:2000, RLM3121, Ruiying Biological), Phospho-Chk1 (Ser345) (1:1000, 2348, Cell Signaling), and Phospho-Chk2 (Thr68) (1:1000, 2197, Cell Signaling). After incubation with corresponding horseradish peroxidase-conjugated secondary antibodies, SuperSignal West Pico Chemiluminescent Substrate (34577, Thermo Fisher Scientific) was used to visualize via chemiluminescence. All blots derive from the same experiment and that they were processed in parallel.
Mouse studies
C57BL/6 J mice were purchased from GemPharmatech Company (Nanjing, China). Animals were housed in standard pathogen-free conditions under 12 h day/night cycle and were allowed ad libitum access to water and a standard diet (XieTong Biotech, Nanjing, China). Eight-week-old female mice were given daily intraperitoneal injections of HU (200 mg/kg of body weight) or a combination of HU and Chk1i (10 mg/kg of body weight) for 8 days. On the fourth day, mice were anesthetized with isoflurane and subjected to 70% partial hepatectomy. The weight of regenerated livers as well as the whole body was measured at 4 days post-PHx. The ratio of wet weight of the regenerated liver remnants after PHx over the weight of the whole animal was taken as the liver-to-body weight ratio. At the end of the animal experiment, mice were anesthetized to collect blood and liver tissue and then euthanized.
For serum analysis, blood was allowed to coagulate for 2 h at 4 °C and serum was collected after centrifugation at 2000 rpm for 10 min at 4 °C and stored at −80 °C. Hepatic function was analyzed by commercial kits of aspartate aminotransferase (ab263882, Abcam, UK), alanine transaminase (RDR-ALT-Mu, Reddot Biotech, Canada), and albumin (E-90AL, Immunology Consultants Laboratory, USA) according to the manufacturer’s instructions. Data were acquired using a Spark microplate reader (Tecan, Switzerland).
For immunofluorescence staining, the liver tissues were fixed in 4% PFA for 1 h and transferred to PBS, then to 30% sucrose for 1 day, embedded in O.C.T. compound (Tissue Tek, Sakura) and sectioned at a thickness of 10 µm using a cryomicrotome (CM 1950, Leica). Antigen retrieval was performed for 10 min in Tris/EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, 0.05% Tween-20, pH 9.0) at 95 °C. Cell apoptosis was measured using the In Situ Cell Death Detection Kit (Roche) following the manufacturer’s instructions. Then sections were stained with primary antibody against HNF4α (1:50, SC6556, Santa Cruz, Dallas, TX) and compatible fluorescently labeled secondary antibodies (Invitrogen, 1:500 dilution).
Quantification and statistical analysis
Unpaired two-tailed Student’s t-test was used for statistical analysis by GraphPad Prism 8; P < 0.05 was considered statistically significant. Quantitative data were shown as means ± SEM.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Lin, S. et al. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 556, 244–248 (2018).
Michalopoulos, G. K. Liver regeneration: alternative epithelial pathways. Int. J. Biochem. Cell Biol. 43, 173–179 (2011).
Riehle, K. J., Dan, Y. Y., Campbell, J. S. & Fausto, N. New concepts in liver regeneration. J. Gastroenterol. Hepatol. 26, 203–212 (2011).
Duncan, A. W., Dorrell, C. & Grompe, M. Stem cells and liver regeneration. Gastroenterology 137, 466–481 (2009).
Miyajima, A., Tanaka, M. & Itoh, T. Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14, 561–574 (2014).
Ko, S., Russell, J. O., Molina, L. M. & Monga, S. P. Liver progenitors and adult cell plasticity in hepatic injury and repair: knowns and unknowns. Annu. Rev. Pathol. 15, 23–50 (2020).
Weng, H.-L. et al. Two sides of one coin: massive hepatic necrosis and progenitor cell-mediated regeneration in acute liver failure. Front. Physiol. 6, 178 (2015).
He, J., Lu, H., Zou, Q. & Luo, L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in Zebrafish. Gastroenterology 146, 789–800 (2014).
Choi, T. Y., Ninov, N., Stainier, D. Y. R. & Shin, D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology 146, 776–788 (2014).
Lu, W.-Y. et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 17, 971–983 (2015).
Raven, A. et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 547, 350–354 (2017).
Deng, X. et al. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell 23, 114–122 (2018).
Russell, J. O. et al. Hepatocyte-specific beta-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology 69, 742–759 (2019).
He, J. et al. Mammalian target of rapamycin complex 1 signaling is required for the dedifferentiation from biliary cell to bipotential progenitor cell in zebrafish liver regeneration. Hepatology 70, 2092–2106 (2019).
He, J. et al. DNA methylation maintenance at the p53 locus initiates biliary-mediated liver regeneration. NPJ Regen. Med. 7, 21 (2022).
Cai, P. et al. Farnesoid X receptor is required for the redifferentiation of bipotential progenitor cells during biliary-mediated zebrafish liver regeneration. Hepatology 74, 3345–3361 (2021).
Huang, M. et al. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology 60, 1753–1766 (2014).
Choi, T. Y. et al. Bone morphogenetic protein signaling governs biliary-driven liver regeneration in zebrafish through tbx2b and id2a. Hepatology 66, 1616–1630 (2017).
Ko, S. et al. Hdac1 regulates differentiation of bipotent liver progenitor cells during regeneration via Sox9b and Cdk8. Gastroenterology 156, 187–202 (2019).
Lavin, M. F. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26, 7749–7758 (2007).
Syed, A. & Tainer, J. A. The MRE11–RAD50–NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 87, 263–294 (2018).
Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Roos, W. P., Thomas, A. D. & Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 16, 20–33 (2016).
Carten, J. D., Bradford, M. K. & Farber, S. A. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 360, 276–285 (2011).
Wilda, M., Demuth, I., Concannon, P., Sperling, K. & Hameister, H. Expression pattern of the Nijmegen breakage syndrome gene, Nbs1, during murine development. Hum. Mol. Genet. 9, 1739–1744 (2000).
Carney, J. P. et al. The hMre11/hRad50 protein complex and nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93, 477–486 (1998).
Aravind, L., Walker, D. R. & Koonin, E. V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27, 1223–1242 (1999).
Petrini, J. H. J., Bressan, D. A. & Yao, M. S. The RAD52 epistasis group in mammalian double strand break repair. Semin. Immunol. 9, 181–188 (1997).
Hoffman, E. A., McCulley, A., Haarer, B., Arnak, R. & Feng, W. Break-seq reveals hydroxyurea-induced chromosome fragility as a result of unscheduled conflict between DNA replication and transcription. Genome Res. 25, 402–412 (2015).
Unno, J. et al. Artemis-dependent DNA double-strand break formation at stalled replication forks. Cancer Sci 104, 703–710 (2013).
Stewart, G. S. et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99, 577–587 (1999).
Zhu, J., Petersen, S., Tessarollo, L. & Nussenzweig, A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 11, 105–109 (2001).
Xiao, Y. & Weaver, D. T. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 25, 2985–2991 (1997).
Luo, G. et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 96, 7376–7381 (1999).
Friedberg, E. C. DNA damage and repair. Nature 421, 436–440 (2003).
Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K. & Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 (2004).
Falck, J., Coates, J. & Jackson, S. P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611 (2005).
Harper, J. W. & Elledge, S. J. The DNA damage response: ten years after. Mol. Cell 28, 739–745 (2007).
Kruiswijk, F., Labuschagne, C. F. & Vousden, K. H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–405 (2015).
Oren, M. Decision making by p53: life, death and cancer. Cell Death Differ. 10, 431–442 (2003).
Hafner, A., Bulyk, M. L., Jambhekar, A. & Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 20, 199–210 (2019).
Berghmans, S. et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl Acad. Sci. USA 102, 407–412 (2005).
Symington, L. S. & Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011).
Jazayeri, A. et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8, 37–45 (2006).
Bakr, A. et al. Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nucleic Acids Res. 43, 3154–3166 (2015).
Zeman, M. K. & Cimprich, K. A. Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014).
Varon, R. et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in nijmegen breakage syndrome. Cell 93, 467–476 (1998).
Taylor, A. M. R. et al. Chromosome instability syndromes. Nat. Rev. Dis. Primers 5, 64 (2019).
Waltes, R. et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am. J. Hum. Genet. 84, 605–616 (2009).
Buis, J. et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135, 85–96 (2008).
Shin, U. et al. Large-scale generation and phenotypic characterization of zebrafish CRISPR mutants of DNA repair genes. DNA Repair 107, 103173 (2021).
Reaper, P. M. et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 7, 428–430 (2011).
Mateos-Gomez, P. A. et al. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015).
Zhang, J. et al. Tel2 regulates redifferentiation of bipotential progenitor cells via Hhex during zebrafish liver regeneration. Cell Rep. 39, 110596 (2022).
Cai, P., Mao, X., Zhao, J. & Luo, L. Ribosome biogenesis protein Urb2 regulates hematopoietic stem cells development via P53 pathway in zebrafish. Biochem. Biophys. Res. Commun. 497, 776–782 (2018).
Zhang, W. et al. Formimidoyltransferase cyclodeaminase prevents the starvation-induced liver hepatomegaly and dysfunction through downregulating mTORC1. PLoS Genet. 17, e1009980 (2021).
Chang, N. et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472 (2013).
Liu, C. et al. Macrophages mediate the repair of brain vascular rupture through direct physical adhesion and mechanical traction. Immunity 44, 1162–1176 (2016).
Yang, Y. et al. A single-cell-resolution fate map of endoderm reveals demarcation of pancreatic progenitors by cell cycle. Proc. Natl Acad. Sci. USA 118, e2025793118 (2021).
Chen, J. et al. Cerebrovascular injuries induce lymphatic invasion into brain parenchyma to guide vascular regeneration in Zebrafish. Dev. Cell 49, 697–710 (2019).
Lu, H., Ma, J., Yang, Y., Shi, W. & Luo, L. EpCAM is an endoderm-specific Wnt derepressor that licenses hepatic development. Dev. Cell 24, 543–553 (2013).
Yang, Y. et al. Intestinal precursors avoid being misinduced to liver cells by activating Cdx-Wnt inhibition cascade. Proc. Natl Acad. Sci. USA 119, e2205110119 (2022).
Chen, J. et al. Acute brain vascular regeneration occurs via lymphatic transdifferentiation. Dev. Cell 56, 3115–3127 (2021).
Acknowledgements
We thank Honghui Huang for providing the p53 mutant and our lab members for helpful discussions and insights about this work. This work was supported by the National Natural Science Foundation of China (32122033, 32192400, 31970784), the National Key R&D Program of China (2021YFA0805000, 2019YFA0802703, 2017YFA0106600), and the Innovation Research 2035 Pilot Plan of Southwest University (SWU-XDPY22008).
Author information
Authors and Affiliations
Contributions
J.H., R.N., and L.L. (Lingfei Luo) designed the experimental strategy, analyzed data, and wrote the manuscript; X.L. and Z.H. performed mice experiment; L.L. (Lianghui Li) performed p53 mutant experiment; L.L. (Linke Li) performed the BODIPY assay; J.S. and J.M. performed all the other experiments in the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, J., Ma, J., Liu, X. et al. The MRN complex maintains the biliary-derived hepatocytes in liver regeneration through ATR-Chk1 pathway. npj Regen Med 8, 20 (2023). https://doi.org/10.1038/s41536-023-00294-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41536-023-00294-3
This article is cited by
-
Harnessing metabolism of hepatic macrophages to aid liver regeneration
Cell Death & Disease (2023)