Abstract
Quantum state tomography is a daunting challenge of experimental quantum computing, even in moderate system size. One way to boost the efficiency of state tomography is via local measurements on reduced density matrices, but the reconstruction of the full state thereafter is hard. Here, we present a machine-learning method to recover the ground states of \(k\)-local Hamiltonians from just the local information, where a fully connected neural network is built to fulfill the task with up to seven qubits. In particular, we test the neural network model with a practical dataset, that in a 4-qubit nuclear magnetic resonance system our method yields global states via the 2-local information with high accuracy. Our work paves the way towards scalable state tomography in large quantum systems.
Similar content being viewed by others
Introduction
Quantum state tomography (QST) plays a vital role in validating and benchmarking quantum devices,1,2,3,4,5 because it can completely capture properties of an arbitrary quantum state. However, QST is not feasible for large systems because of its need for exponential resources. In recent years, there has been extensive research on methods for boosting the efficiency of QST.6,7,8,9,10,11,12 One of the promising candidates among these methods is QST via reduced density matrices (RDMs);13,14,15,16,17,18,19 because local measurements are convenient and accurate on many experimental platforms.
QST via RDMs is also a useful tool for characterizing ground states of local Hamiltonians. A many-body Hamiltonian \(H\) is \(k\)-local if \(H={\sum }_{i}{H}_{i}^{(k)}\), where each term \({H}_{i}^{(k)}\) acts non-trivially on at most \(k\) particles. For \(k\)-local Hamiltonians, only polynomial number of parameters are needed to characterize the whole system. Moreover, generally, a single eigenstate of such \(k\)-local Hamiltonian can encode the information of the system.18,20,21 Therefore, for these ground states, one only needs \(k\)-local measurements for state tomography. Although local measurements are efficient and even if \(\left|\psi \right\rangle\) is uniquely determined by its \(k\)-local measurements, reconstructing \(\left|\psi \right\rangle\) from its \(k\)-local measurements is computationally hard.22 We remark that this is not due to the problem that \(\left|\psi \right\rangle\) needs to be described by exponentially many parameters. In fact, in many cases, ground states of \(k\)-local Hamiltonians can be effectively represented by tensor product states.18,23
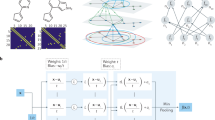
The state reconstruction problem naturally connects to the regression problem in supervised learning. Regression analysis, in general, seeks to discover the relation between inputs and outputs, i.e., to recover the underlying mathematical model. Unsupervised learning techniques have been applied to QST in various cases, such as in refs. 24,25 In our case, as shown in Fig. 1, by knowing the Hamiltonian \(H\), it is relatively easy to get the ground state \(\left|{\psi }_{H}\right\rangle\) since the ground state is nothing but the eigenvector corresponding to the smallest eigenvalue. And then we could naturally achieve the \(k\)-local measurements \({\bf{M}}\) of \(\left|{\psi }_{H}\right\rangle\). Therefore, the data for tuning our reverse engineering model is accessible, which allows us to realize QST through supervised learning practically. Additionally, artificial neural networks are often noise tolerable,26,27,28 so they are favorable for working with experimental data.
Procedure of our neural network based local quantum state tomography method. As shown by the dashed arrows, we first construct training and test dataset by generating random \(k\)-local Hamiltonians \(H\), calculate their ground states \(\left|{\psi }_{H}\right\rangle\), and obtain local measurement results \({\bf{M}}\). We then train the neural network with the generated training dataset. After training, as represented by the black arrows, we first obtain the Hamiltonian \(H\) through local measurement results \({\bf{M}}\) from the neural network, then recover the ground states from the obtained Hamiltonian. In contrast, the red arrow represents the direction of the normal QST process, which is computationally hard.
In this work, we propose a local-measurement-based QST by fully connected feedforward neural network, in which every neuron connects to every neuron in the next layer and information only passes forward (i.e., there is no loop in the network). We first build a fully connected feedforward neural network for \(4\)-qubit ground states of fully connected \(2\)-local Hamiltonians. Our trained \(4\)-qubit network not only analyzes the test dataset with high fidelity but also reconstruct \(4\)-qubit nuclear magnetic resonance (NMR) experimental states accurately. We use the \(4\)-qubit case to demonstrate the potential of using neural networks to realize QST via \(k\)-local measurements. The versatile framework of neural networks for recovering ground states of \(k\)-local Hamiltonians could be extended to more qubits and various interaction structures; we then apply our methods to the ground states of seven-qubit 2-local Hamiltonians with nearest-neighbor couplings. In both cases, neural networks give accurate estimates with high fidelities. We observe our framework yields higher efficiency and better noise tolerance compared with the least-squares tomography (the approximated maximum likelihood estimation (MLE)) when the added noise is >5%.
Results
Theory
The universal approximation theorem29 states that every continuous function on the compact subsets of \({{\mathbb{R}}}^{n}\) can be approximated by a multi-layer feedforward neural network with a finite number of neurons, i.e., computational units. And by observing the relation between \(k\)-local Hamiltonian and local measurements of its ground state, as shown in Fig. 1, we are empowered to turn the tomography problem to a regression problem which fits perfectly into the neural network framework.
In particular, we first construct a deep neural network for \(4\)-qubit ground states of full \(2\)-local Hamiltonians as follows:
where \({\sigma }_{k},{\sigma }_{n},{\sigma }_{m}\in \Delta\), and \(\Delta =\{{\sigma }_{1}={\sigma }_{x},{\sigma }_{2}={\sigma }_{y},{\sigma }_{3}={\sigma }_{z},{\sigma }_{4}=I\}\).
We denote the set of Hamiltonian coefficients as \(\overrightarrow{h}=\{{\omega }_{k}^{(i)},{J}_{nm}^{(ij)}\}\). The coefficient vector \(\overrightarrow{h}\) is the vector representation of \(H\) according to the basis set \({\bf{B}}=\{{\sigma }_{m}\otimes {\sigma }_{n}:n+m\,\ne \,8,{\sigma }_{m},{\sigma }_{n}\in \Delta \}\). The configuration of the ground states is illustrated in Fig. 2a.
The number of parameters of the local observables of ground states determines the amount of input units of the neural network. Concretely, \({\bf{M}}=\{{s}_{m,n}^{(i,j)}:{s}_{m,n}^{(i,j)}={\rm{Tr}}({{\rm{Tr}}}_{(i,j)}\rho \cdot {B}_{(m,n)}),{B}_{(m,n)}\in {\bf{B}},1\le i\;<\;j\le 4,1\le n,m\le 4\}\), where \({\sigma }_{n},{\sigma }_{m}\in \Delta\) and \(\rho\) is the density matrix of the ground state. \({\bf{M}}\) is a set of true expectation values \({s}_{m,n}^{(i,j)}\) of the local observables \({B}_{(m,n)}\) in the ground states \(\rho\). Notice that we are using the true values of expectation values instead of their estimations (which contain statistical fluctuations), since we are theoretically generating all the training and testing data. The input layer has \(66\) neurons since the cardinality of the set of measurement results is \(66\). Our network then contains two fully connected hidden layers, in which every neuron in the previous layer is connected to every neuron in the next layer. The number of output units equals to the number of parameters of our \(2\)-local Hamiltonian, which is \(66\) in our \(4\)-qubit case. More details of our neural network can be found in “Methods” section.
Our training data consist of the 120,000 randomly generated \(2\)-local Hamiltonians as output and the local measurements of their corresponding ground states. The test data include 5000 pairs of Hamiltonians and local measurement results \(({H}_{i},{{\bf{M}}}_{i})\).
We train the network by a popular optimizer in the machine-learning community called Adam (adaptive moment estimation).30,31 For loss function, we choose cosine proximity \(\cos (\theta )=({\overrightarrow{h}}_{{\rm{pred}}}\cdot \overrightarrow{h})/(\parallel {\overrightarrow{h}}_{{\rm{pred}}}\parallel \cdot \parallel \overrightarrow{h}\parallel )\), where \({\overrightarrow{h}}_{{\rm{pred}}}\) is the estimate of the neural network and \(\overrightarrow{h}\) is the desired output. Generally speaking, the role of loss functions in supervised learning is to efficiently measure the distance between the true value and the estimated outcome. (In our case, it is the distance between \(\overrightarrow{h}\) and \({\overrightarrow{h}}_{{\rm{pred}}}\)). And the training procedure seeks to minimize this distance. We find the cosine proximity function fits our scenario better than the more commonly chosen loss functions, such as mean square error or mean absolute error.32 The reason can be understood as follows. Because the parameter vector \(\overrightarrow{h}\) is a representation of the corresponding Hamiltonian in the Hilbert space expanded by the local operators \({\bf{B}}\), the angle \(\theta\) between the two vectors \(\overrightarrow{h}\) and \({\overrightarrow{h}}_{{\rm{pred}}}\) is a “directional distance measure” between two corresponding Hamiltonians.20 Notice that the Hamiltonian corresponding to the parameter \(\overrightarrow{h}\) has the same eigenvectors as those of the Hamiltonian of \(c\cdot \overrightarrow{h}\), where \(c\in {\mathbb{R}}\) is a constant. In other words, we only care about the “directional distance”. Instead of forcing every single element close to its true value (as mean squared error or mean absolute error does), the cosine loss function tends to train the angle \(\theta\) towards zero, which is more adapted to our situation.
As illustrated in Fig. 1, after getting estimated Hamiltonian from the neural network, we calculate the ground state \({\rho }_{{\rm{nn}}}\) of the estimated Hamiltonian and take the result as the estimate of the ground state that we attempt to recover. We remark that our estimated Hamiltonian is not necessarily exactly the same as the original Hamiltonian; Even if that happens, our numeric results suggest their ground states are still close.
There are two different fidelity functions that we can use to measure the distance between the randomly generated states \({\rho }_{{\rm{rd}}}\) and our neural network estimated states \({\rho }_{{\rm{nn}}}\), namley:
The fidelity measure \(f\) defined in Eq. (2) is standard,33 which requires the matrix \({\rho }_{1}\) and \({\rho }_{2}\) to be positive semi-definite. Considering that the density matrix obtained directly from the raw data of a state tomography experiment may possibly not be positive definite, we usually adopt the definition of \(C\) for processing the raw data in NMR.34 In this work, there are not negative matrices after constraining the raw density matrices to be positive semi-definite. The values of the fidelities are calculated by \(f\) if there is no additional explanation in the following.
After supervised learning on the training data, our neural network is capable of estimating the 4-qubit output of the test set with high performance. The fidelity averaged over the whole test set is 98.7%. The maximum, minimum, standard deviations of fidelities for the test set are shown in Table 1. Figure 2c illustrates the fidelities between 100 random states \({\rho }_{{\rm{rd}}}\) and our neural network estimates \({\rho }_{{\rm{nn}}}\).
Theoretical results for 4 qubits and 7 qubits. a The configuration of our 4-qubit states. Each dot presents a qubit, and every qubit interacts with each other. b The configuration of our 7-qubit states: only nearest qubits have interactions. c The fidelities of 100 random 4-qubit states \({\rho }_{{\rm{rd}}}\) and our neural network estimates \({\rho }_{{\rm{nn}}}\). Notice that the \(x\)-coordinate does not have physical meaning, we randomly pick 100 states and label them from \(0\) to \(99\). It is the same for the 7-qubit case. The average fidelity (Eq. (2)) is \(98.7 \%\). d The fidelities of 100 random 7-qubit states \({\rho }_{{\rm{rd}}}\) and our neural network estimates \({\rho }_{{\rm{nn}}}\). The average fidelity (Eq. (2)) of the whole test data set is \(97.9 \%\).
Our framework generalizes directly to more qubits and different interaction patterns. We apply our framework to recover 7-qubit ground states of \(2\)-local Hamiltonians with nearest-neighbor interaction. The configuration of our \(7\)-qubit states is shown in Fig. 2b. The Hamiltonian of this 7-qubit case is
where \({\sigma }_{k},{\sigma }_{n},{\sigma }_{m}\in \Delta\), \({\omega }_{k}^{(i)}\) and \({J}_{nm}^{(i)}\) are coefficients. We trained a similar neural network with 250,000 pairs of randomly generated Hamiltonians and \(2\)-local measurements of the corresponding ground states. For the 5000 randomly generated test sets, the network estimates have an average fidelity of 97.9%. More statistical performance are shown in Table 1 and fidelity results of 100 random generated states are shown in Fig. 2d.
Due to the variance inherent to this method, it is natural to ask how to determine whether a neural network estimate \({\rho }_{{\rm{nn}}}\) is acceptable without knowing the true state \({\rho }_{{\rm{rd}}}\). This problem can be easily solved by calculating the measurement estimate \({{\bf{M}}}_{{\rm{pred}}}\), i.e., using the estimate \({\rho }_{{\rm{nn}}}\) to measure the set of local operators \({\bf{B}}\). By setting an acceptable error bound and comparing \({{\bf{M}}}_{{\rm{pred}}}\) with the true measurements \({\bf{M}}\), one can decide whether to accept \({\rho }_{{\rm{nn}}}\) or not. Please see the “Methods” section for details.
Our neural-network-based framework is also significantly faster than the approximated MLE method. Once the network is trained sufficiently well, it can be used to deal with thousands of datasets without much effort on a regular computer. Calculating \({\rho }_{{\rm{nn}}}\) from \({\overrightarrow{h}}_{{\rm{pred}}}\), which is essentially the computation of the eigenvector corresponding to the smallest eigenvalue, is the only part that may take some time. Detailed discussions could be found in the “Methods” section.
Experiment
So far, our theoretical model is noise-free. To demonstrate that our trained machine-learning model is resilient to experimental noises, we experimentally prepare the ground states of the random Hamiltonians and then try to reconstruct the final quantum states from 2-local measurements using a four-qubit NMR platform.35,36,37,38 The four-qubit sample is 13C-labeled trans-crotonic acid dissolved in d6-acetone, where C1–C4 are encoded as the four work qubits, and the rest spin-half-nuclei are decoupled throughout all experiments. Figure 3 describes the parameters and structure of this molecule. Under the weak-coupling approximation, the Hamiltonian of the system writes
where \({\nu }_{j}\) are the chemical shifts, \({J}_{jk}\) are the J-coupling strengths, and \({\nu }_{0}\) is the reference frequency of 13C channel in the NMR platform. All experiments were carried out on a Bruker AVANCE 400 MHz spectrometer at room temperature. We briefly describe our three experimental steps here and leave the details in the “Methods” section: (i) Initialization: The pseudo-pure state39,40,41 for being the input of quantum computation \(\left|0000\right\rangle\) is prepared. (More details are provided in the “Methods” section). (ii) Evolution: Starting from the state \(\left|0000\right\rangle\), we create the ground state of the random two-body Hamiltonian by applying the optimized shaped pulses. (iii) Measurement: In NMR experiments, the expectation values of all 2-qubit Pauli products can be measured by the ensemble measurement. From them, we can directly obtain all 2-local measurements, and perform four-qubit QST to estimate the quality of our implementations, which is accomplished by the least-squares tomography from the experimental data. More details about the least-squares tomography can be found in “Methods” section.
The molecular structure and Hamiltonian parameters of the 13C-labeled trans-crotonic acid. The atoms C1–C4 are used as the four qubits in the experiment, and the atoms M, H1 and H2 are decoupled throughout the experiment. In the table, the chemical shifts with respect to the Larmor frequency and J-coupling constants (in Hz) are listed by the diagonal and off-diagonal numbers, respectively. The relaxation timescales \({T}_{2}\) (in seconds) are shown at the bottom.
In experiments, we created the ground states of 20 random Hamiltonians of the form in Eq. (1) and performed 4-qubit QST for them after the state preparations. It is worth emphasizing that the experimental raw density matrices obtained from ensemble measurements on NMR are usually negative. First, we further performed the least-squares QST from the raw density matrices in experiments as \({\rho }_{\exp }\), and estimated that the fidelities between the experimental states \({\rho }_{\exp }\) and the target ground state \({\rho }_{{\rm{th}}}\) are over \(99.2\)%. It is noted that the purpose of reconstructing the states \({\rho }_{\exp }\) is to use them to compare with the results estimated by our neural network. We collected the expectation values of all 2-qubit Pauli product operators, such as \(\left\langle {\sigma }_{x}\otimes I\otimes I\otimes I\right\rangle\) and \(\left\langle {\sigma }_{x}\otimes {\sigma }_{y}\otimes I\otimes I\right\rangle\), which were directly obtained by measuring the expectation values of these Pauli strings in NMR. Then we fed them into our neural-network-based framework to reconstruct the 4-qubit states, obtaining an average fidelity of 98.8% between \({\rho }_{\exp }\) and \({\rho }_{{\rm{nn}}}\), where \({\rho }_{{\rm{nn}}}\) is the neural network estimated state. Figure 4 shows the fidelity details of these density matrices. The results indicate that the original 4-qubit state can be efficiently reconstructed by our trained neural network using only 2-local measurements, instead of the traditional full QST.
The predication results with experimental data. Here we list three different fidelities for 20 experimental instances. The horizontal axis is the dummy label of the 20 experimental states. The cyan bars, \({f}_{\exp \!-{\rm{th}}}\), are the fidelities between the theoretical states \({\rho }_{{\rm{th}}}\) and the experimental states \({\rho }_{\exp }\). The blue triangles, \({f}_{\exp \!-{\rm{nn}}}\), are fidelities between our neural network estimates \({\rho }_{{\rm{nn}}}\) and the experimental states \({\rho }_{\exp }\) with the average fidelity over 98.8%. And the green dots, \({f}_{{\rm{nn}}\!-{\rm{th}}}\), are the fidelities between our neural network estimates and the theoretical states.
Discussion
As a famous double-edged sword in experimental quantum computing, QST captures full information of quantum states on the one hand, while on the other hand, its implementation consumes a tremendous amount of resources. Unlike traditional QST that requires exponentially many experiments with the growth of system size, the recent approach by measuring RDMs and reconstructing the full state thereafter opens up a new avenue to efficiently realize experimental QST. However, there is still an obstacle in this approach, that it is in general computationally hard to construct the full quantum state from its local information.
This is a typical problem empowered by machine learning. In this work, we apply the neural network model to solve this problem and demonstrate the feasibility of our method with up to seven qubits in the simulation. It should be noticed that 7-qubit QST in experiments is already a significant challenge in many platforms—the largest QST to date is of 10 qubits in superconducting circuits, where the theoretical state is a GHZ state with rather simple mathematical form.61 We further demonstrate that our method works well in a 4-qubit NMR experiment, thus validating its usefulness in practice. We anticipate this method to be a powerful tool in future QST tasks of many qubits due to its accuracy and convenience. Comparing with the MLE, our method has acceptable fidelities, better noise tolerance and also has a significant advantage in terms of speed.
Our framework can be extended in several ways. First, we can consider excited states. As stated in the “Results” section, the Hamiltonian recovered by our neural network is not necessarily the original Hamiltonian, but their ground states are fairly close. We preliminarily examined the eigenstates of estimated Hamiltonians. Although the ground states have considerable overlap, the excited states are not close to each other. It means, in this reverse engineering problem, ground states are numerically more stable than excited states. To recover excited states using our method, one may need to use more sophisticated neural networks, such as convolutional neural network62 (CNN) or residual neural network63 (ResNet). Second, although we have not included noise in the training and test data, our network estimates the experimental 4-qubit fully connected 2-local states with high fidelities. This indicates our method has certain error tolerant ability. For future study, one can add different noise to the training and test data. Third, one can also study how to incorporate the current method into the existing quantum tomography methods, such as compressive sensing techniques.9,64,65
Methods
Machine learning
In this subsection, we discuss our training/test dataset generation procedure, the structure, and hyperparameters of our neural network, and the required number of training data during training. And we also provide a criterion for determining whether the neural network estimate is acceptable without knowing the true states.
The training and test data sets are formed by random \(k\)-local Hamiltonians and \(k\)-local measurements of corresponding ground states. For our 4-qubit case, the 2-local Hamiltonians are defined in Eq. (1). The parameter vector \(\overrightarrow{h}\) of random Hamiltonians are uniformly drawn from random normal distributions without uniform mean values and standard deviations. It is realized by applying function np.random.normal in Python. Similarly, for the 7-qubit case, Hamiltonian is defined in Eq. (4), and the corresponding parameter vector \(\overrightarrow{h}\) is generated by the same method. As the dashed lines in Fig. 1 shows, after getting random Hamiltonians \(H\), we calculate the ground states \(\left|{\psi }_{H}\right\rangle\) (the eigenvector corresponds to the smallest eigenvalue of \(H\)) and then get the 2-local measurements \({\bf{M}}\).
In this work, we use a fully connected feedforward neural network, which is famous as the first and most simple type of neural network.42 By fully connected, it means every neuron is connected to every other neuron in the next layer. Feedforward or acyclic, as the word indicated, means information only passes forward; the network has no cycle. Our machine-learning process is implemented using Keras,43 which is a high-level deep learning library running on top of the popular machine-learning framework: Tensorflow.44
As mentioned in the “Results” section, the true value of local measurements have been used as input to our neural network. The input is \({\bf{M}}=\{{s}_{m,n}^{(i,j)}:{s}_{m,n}^{(i,j)}={\rm{Tr}}({{\rm{Tr}}}_{(i,j)}\rho \cdot {B}_{(m,n)}),{B}_{(m,n)}\in {\bf{B}},1\le i<j\le 4,1\le n,m\le 4\}\). For the 4-qubit case, it is easy to see that \({\bf{M}}\) has \(3\,\times\, 4=12\) single body terms and \({C}_{4}^{2}\times 9=54\) 2-body terms. By arranging these \(66\) elements in \({\bf{M}}\) into a row, we set it as the input of our neural network.
The output set to be the vector representation of the Hamiltonian \(\overrightarrow{h}\), which also has 66 entries. For the 7-qubit 2-local case, where 2-body terms only appear on nearest qubits, the network takes 2-local measurements as input, and the number of neurons in the input layer is \(7\,\times\, 3+6\,\times\, 3\,\times\, 3=75\). The number of neurons in the output layer is also 75.
The physical aspect of our problem fixes the input and output layers. The principle for choosing the number of hidden layers is efficiency. While training networks, inspired by Occam’s Razor principle, we choose fewer layers and neurons when increasing them do not significantly increase the performance but increases the required training epochs. In our 4-qubit case, two hidden layers of 300 neurons have been inserted between the input layer and the output layer. In the 7-qubit case, we use four fully connected hidden layers with the following number of hidden neurons: 150-300-300-150. The activate function for each layer is rectified linear unit (ReLU),45 which is a widely used non-linear activation function. We also choose the optimizer having the best performance in our problem over almost all the built-in optimizers in Tensorflow: AdamOptimizer (adaptive moment estimation).30 The learning rate is set to be 0.001.
The whole training dataset has been split into two parts, 80% used for training, and 20% used for validation after each epoch. A new data set of 5000 data was used as the test set after training. The initial batch size was chosen as 512. As the amount of training data increases, the average fidelity of estimated states and the true test states goes up. The neural network reaches a certain performance after we fed sufficient training data. More training data requires more training epochs; however, replete epochs ebb the neural network performance due to over-fitting. Table 2 shows the average fidelities of using different training data and epochs. The first round of training locks down the optimal amount of training data, then we change the batch size and find the optimal epoch. We report the results for the second round training in Table 3. For the 4-qubit case, appropriate increase in the batch size can benefit the stability of training process, thus improves the performance of the neural network. Though, by choosing the batch size as 512 and 2048, the network can also reach the same performance with larger epochs, we chose the batch size as 1028, since more epochs require more training time. After the same attempting for the 7-qubit case, we find 512 a promising batch size.
The time cost for preparing the network involves two parts—generating training and testing data, and training the networks. Most of the time spending on data generating is to solve the ground states (eigenvector corresponding to the smallest eigenvalue) of randomly generated Hamiltonians. It takes roughly 5 min (2.2 h) to generate the whole data set for 4-qubit (7-qubit) by implementing eigs in MATLAB. With sufficient data in hand, the network-training procedure takes about 12 min (49 min) for 4-qubit (7-qubit).
As reported in the “Results” section, the fidelities of neural network estimates have a slight variation, for example, the fidelity of the 4-qubit case is range from 91.4% to 99.8%. One who uses this framework might wonder how precise the neural network outcome is compared to the true state. In contrast of the scenario when we are testing our framework theoretically, we do not have the true state in hand. Now it is natural to ask that how to determine whether the estimate is precise enough. Providentially, we could solve this question in a straightforward way.
Based on \({\rho }_{{\rm{nn}}}\), we compute \({{\bf{M}}}_{{\rm{pred}}}=\{{s}_{m,n}^{(i,j)}:{s}_{m,n}^{(i,j)}={\rm{Tr}}({{\rm{Tr}}}_{(i,j)}{\rho }_{{\rm{nn}}}\cdot {B}_{(m,n)}),{B}_{(m,n)}\in {\bf{B}},1\le i<j\le 4,1\le n,m\le 4\}\) and compare with the original \({\bf{M}}\). Root-mean-square-error (RMSE) between two variables \(\overrightarrow{x}\) and \(\overrightarrow{y}\), defined as \({\rm{rmse}}(\overrightarrow{x},\overrightarrow{y})=\sqrt{\frac{1}{d}{\sum }_{i=1}^{d}{({x}_{i}-{y}_{i})}^{2}}\), is a frequently used quantity to measure the closeness of \(\overrightarrow{x}\) and \(\overrightarrow{y}\). In reality, how bad an error is also depends on the magnitude of the true value. That means, with the same RMSE, the larger the magnitude of the true value is the better the accuracy. A measure referring to the real value reveals more about how precise between an estimation and a expected outcome. We, therefore, define a quantity called relative RMSE, namely \({\rm{rrmse}}(\overrightarrow{x},\overrightarrow{y})=\sqrt{\frac{1}{d}{\sum }_{i=1}^{d}{({x}_{i}-{y}_{i})}^{2}}/| | \overrightarrow{y}| | ={\rm{rmse}}(\overrightarrow{x},\overrightarrow{y})/| | \overrightarrow{y}| |\), where \(y\) is the true value and \(| | \overrightarrow{y}| |\) is its \({l}^{2}\)-norm. The relative RMSE between \({{\bf{M}}}_{{\rm{pred}}}\) and \({\bf{M}}\) is \({\rm{rmse}}({{\bf{M}}}_{{\rm{pred}}},{\bf{M}})/| | {\bf{M}}| |\). By bounding the relative RMSE <0.2%, 4692 out of 5000 (93.8%) estimations of our 4-qubit network are acceptable, and the probability of these estimations having fidelities higher than 97% is 99.8%.
Comparison with the approximated MLE
The standard MLE46,47,48,49 is usually adopted to reconstruct a legal and full quantum state whose local information is closest to the measured results. It technically maximizes the likelihood of the estimate by the given data. When we make the Gaussian distribution and assume that all measurements have the same standard deviation, the MLE is approximately the least-squares tomography which minimizes the distance between the searched results and the measurement outcomes.50 In this section, we make a comparison of efficiency, accuracy, and noise tolerance between the approximated MLE and our method.
With a personal computer,51 every single 4-qubit state takes about 1 min to compute for the approximated MLE. The estimating procedure of our method analyzed 5000 data in 2 min (about 0.024 s per data set) using the same computer. For the 7-qubit case, the approximated MLE requires about 168 min to converge for each single data point. Remarkably, our method can process 5000 data sets within <6 min (about 0.070 s per data set). This suggests that our method is substantially faster than the approximated MLE. We can reasonably expect that when the system size gets even larger, our computation time advantage will become more impressive.
In the 4-qubit cases, the approximated MLE can yield estimates with an average fidelity of 99.9%. In the 7-qubit cases, it can still achieve an average fidelity of 99.9%. Therefore in terms of accuracy, the approximated MLE slightly outperforms our method.
We also analyze the noise tolerance of the two methods by adding noise to the input measurements. The set of unbiased noise \(\overrightarrow{n}\) was generated according to the normal distribution with mean value \(0\) and standard deviation \(1\). The percentile noise vector \(\alpha \overrightarrow{n}\) is formed by multiplying the factor \(\alpha \in \{5 \% ,10 \% ,15 \% ,20 \% ,25 \% ,30 \% \}\) to the unbiased noise \(\overrightarrow{n}\). By adding \(\alpha\overrightarrow{n}\) to the true measurements \({\bf{M}}\), we formed the noisy input \({\bf{M}}+\alpha\overrightarrow{n}\). Suppose the approximated MLE or our neural network estimates the noisy output \({\rho }_{{\rm{noise}}}\). We calculate the fidelities of the estimate \({\rho }_{{\rm{noise}}}\) with the true state \(\rho\) for 100 pairs of 4-qubit data. As depicted in Fig. 5, our method has better noise tolerance than the approximated MLE with the pure state constraint when noise >5% is added to the measurements of a pure state.
Performances of the approximated MLE and our neural network framework under noisy inputs. The dots in the figure are the average fidelities \(f\) between the true state \(\rho\) and the noisy estimates \({\rho }_{{\rm{noise}}}\) corresponding to various percentage of noise. The red circles are estimated by the neural network, and the blue triangles are from the approximated MLE.
NMR states preparation
Our experimental procedure consists of three steps: initialization, evolution, and measurement. In this subsection, we discuss these three steps in details.
-
(i)
Initialization: The computational basis state \({\left|0\right\rangle }^{\otimes n}\) is usually chosen as the input state for quantum computation. Most of the quantum systems do not start from such an input state, so a proper initialization processing is necessary before applying quantum circuits. In NMR, the sample initially stays in the Boltzmann distribution at room temperature,
$${\rho }_{{\rm{thermal}}}={\mathcal{I}}/16+\epsilon ({\sigma }_{z}^{1}+{\sigma }_{z}^{2}+{\sigma }_{z}^{3}+{\sigma }_{z}^{4}),$$where \({\mathcal{I}}\) is the \(16\,\times\, 16\) identity matrix and \(\epsilon \approx 1{0}^{-5}\) is the polarization. We cannot directly use it as the input state for quantum computation, because such a thermal state is a highly mixed state.39,52 We instead create a so-called pseudo-pure state (PPS) from this thermal state by using the spatial averaging technique,39,40,41 which consists of applying local unitary rotations and using \(z\)-gradient fields to destroy the unwanted coherence. The form of the 4-qubit PPS can be written as
$${\rho }_{0000}=(1-\epsilon ^{\prime} ){\mathcal{I}}/16+\epsilon ^{\prime} \left|0000\right\rangle \left\langle 0000\right|.$$Here, although the PPS \({\rho }_{0000}\) is also a highly mixed state, the identity part \({\mathcal{I}}\) neither changes under any unitary operations nor contributes to observable NMR signal. It means that we can focus on the deviated part \(\left|0000\right\rangle \left\langle 0000\right|\) and consider \(\left|0000\right\rangle \left\langle 0000\right|\) as the initial state of our quantum system. Finally, 4-qubit QST was performed to evaluate the quality of our PPS. We found that the fidelity between the perfect pure state \(\left|0000\right\rangle\) and the experimentally measured PPS is about 98.7% by the definition of \(C\) in Eq. (3), where the raw PPS density matrix obtained directly from the experiment is negative. This sets a solid ground for the subsequent experiments.
-
(ii)
Evolution: In this step, we prepared the ground states of the given Hamiltonians using optimized pulses. The form of the considered Hamiltonian is chosen as Eq. (1).
Here, the parameters \({\omega }_{k}^{(i)}\) and \({J}_{nm}^{(ij)}\) mean the chemical shift and the J-coupling strength, respectively. In experiments, we create the ground states of different Hamiltonians by randomly changing the parameter set \(({\omega }_{k}^{(i)},{J}_{nm}^{(ij)})\). For the given Hamiltonian, the gradient ascent pulse engineering (GRAPE) algorithm53,54,55,56 is adapted to optimize a radio-frequency (RF) pulse to realize the dynamical evolution from the initial state \(\left|0000\right\rangle\) to the target ground state. The GRAPE pulses are designed to be robust to the static field distributions and RF inhomogeneity, and the simulated fidelity is over \(0.99\) for each dynamical evolution.
-
(iii)
Measurement: In principle, we only need to measure the 2-local measurements to determine the original 4-qubit Hamiltonian through our trained network. Experimentally, we performed 4-qubit QST, which naturally includes the 2-local measurements after preparing these states,57,58,59 to evaluate the performance of our implementations. Hence, we can estimate the quality of the experimental implementations by computing the fidelity between the target ground state \({\rho }_{{\rm{th}}}=\left|{\psi }_{{\rm{th}}}\right\rangle \left\langle {\psi }_{{\rm{th}}}\right|\) and the experimentally reconstructed density matrix \({\rho }_{\exp }\).60 By reconstructing states \({\rho }_{{\rm{nn}}}\) merely based on the experimental 2-local measurements, the performance of our trained neural network can be evaluated by comparing the experimental states \({\rho }_{\exp }\) with the states \({\rho }_{{\rm{nn}}}\).
Finally, we attempt to evaluate the confidence of the expected results by analyzing the potential error sources in experiments. The infidelity of the experimental density matrix is mainly caused by some unavoidable factors in experiments, including decoherence effects, imperfections of the PPS preparation, and imprecision of the optimized pulses. From a theoretical perspective, we numerically simulate the influence of the optimized pulses and the decoherence effect of our qubits. Then we compare the fidelity computed in this manner with the ideal case to evaluate the quality of the final density matrix. As a numerical result, about 0.2% infidelity was created on average and the 1.2% error is related to the infidelity of the initial state preparation. Additionally, other errors can also contribute to the infidelity, such as imperfections in the readout pulses and spectral fitting.
The approximated MLE
We briefly describe the approximated MLE we used in the numerical simulation. The standard MLE48,49 is also a method to produce satisfactory results in recovering the full states from the experimental measurements. In general, the standard MLE can be divided into three steps.
- (i)
Parameterize a density matrix in a legal way. Here, we describe a pure density matrix by
$$\rho (\overrightarrow{x})=V(\overrightarrow{x}){V}^{\dagger }(\overrightarrow{x})/{\rm{Tr}}(V(\overrightarrow{x}){V}^{\dagger }(\overrightarrow{x})).$$\(V\) is a \({2}^{N}\)-dimensional vector with the parameters \(\overrightarrow{x}\) and the number of qubits \(N\). \(\rho (\overrightarrow{x})\) is a normalized and non-negative definite Hermitian density matrix under such a parameterization.
- (ii)
Construct a likelihood function to be maximized. The measurements calculated from the parameterized density matrix \(\rho (\overrightarrow{x})\) are \({\rm{Tr}}({{\rm{Tr}}}_{(i,j)}\rho (\overrightarrow{x})\cdot {B}_{(m,n)})\) with \({B}_{(m,n)}\in {\bf{B}},1\le i<j\le 4\) and \(1\le n,m\le 4\), and the total probability of \(\rho (\overrightarrow{x})\) yielding the results closely to the true measurements \({\bf{M}}\) can be written as
$$P(\overrightarrow{x})=\frac{1}{{\mathcal{N}}}\prod _{i,j,m,n}\exp \left[-\frac{{\{{\rm{Tr}}({{\rm{Tr}}}_{(i,j)}\rho (\overrightarrow{x})\cdot {B}_{(m,n)})-{s}_{m,n}^{(i,j)}\}}^{2}}{2{({\chi }_{m,n}^{(i,j)})}^{2}}\right],$$where \({\chi }_{m,n}^{(i,j)}\) is the standard deviation of each measurement \({s}_{m,n}^{(i,j)}\) and \({\mathcal{N}}\) is the normalization (Gaussian model). \(P(\overrightarrow{x})\) is the likelihood function we need to maximize. If we assume the standard deviation is the same, the standard MLE is approximately least-squares tomography.50 It is equivalent to minimize the following function:
$${\mathcal{F}}(\overrightarrow{x})=\sum _{i,j,m,n}{\left[{\rm{Tr}}({{\rm{Tr}}}_{(i,j)}\rho (\overrightarrow{x})\cdot {B}_{(m,n)})-{s}_{m,n}^{(i,j)}\right]}^{2}.$$Here, we ignore some constants which do not influence the optimization, e.g., the normalization factor \({\mathcal{N}}\). \({\mathcal{F}}(\overrightarrow{x})\) is the cost function that we minimize with the least-squares tomography.
- (iii)
Minimize the cost function using some techniques. We use the function lsqnonlin of MATLAB with an initial guess and a default setting. It takes a while to optimize a sum of squares like \({\mathcal{F}}(\overrightarrow{x})\). Finally, quantum state \(\rho (\overrightarrow{x})\) can be recovered from the parameters \(\overrightarrow{x}\) when the optimization is finished.
Data availability
The experimental data and the source code that support the findings of this study are available from the corresponding author on reasonable request.
References
D’Ariano, G. M., De Laurentis, M., Paris, M. G., Porzio, A. & Solimeno, S. Quantum tomography as a tool for the characterization of optical devices. J. Opt. B 4, S127 (2002).
Häffner, H. et al. Scalable multiparticle entanglement of trapped ions. Nature 438, 643 (2005).
Leibfried, D. et al. Creation of a six-atom ‘schrödinger cat’ state. Nature 438, 639 (2005).
Lvovsky, A. I. & Raymer, M. G. Continuous-variable optical quantum-state tomography. Rev. Mod. Phys. 81, 299 (2009).
Baur, M. et al. Benchmarking a quantum teleportation protocol in superconducting circuits using tomography and an entanglement witness. Phys. Rev. Lett. 108, 040502 (2012).
Klimov, A., Munoz, C., Fernández, A. & Saavedra, C. Optimal quantum-state reconstruction for cold trapped ions. Phys. Rev. A 77, 060303 (2008).
Hou, Z. et al. Full reconstruction of a 14-qubit state within four hours. New J. Phys. 18, 083036 (2016).
Cramer, M. et al. Efficient quantum state tomography. Nat. Commun. 1, 149 (2010).
Gross, D., Liu, Y.-K., Flammia, S. T., Becker, S. & Eisert, J. Quantum state tomography via compressed sensing. Phys. Rev. Lett. 105, 150401 (2010).
Tóth, G. et al. Permutationally invariant quantum tomography. Phys. Rev. Lett. 105, 250403 (2010).
Li, J. et al. Optimal design of measurement settings for quantum-state-tomography experiments. Phys. Rev. A 96, 032307 (2017).
Lanyon, B. et al. Efficient tomography of a quantum many-body system. Nat. Phys. 13, 1158 (2017).
Baldwin, C. H., Deutsch, I. H. & Kalev, A. Strictly-complete measurements for bounded-rank quantum-state tomography. Phys. Rev. A 93(5), 052105 (2016). https://journals.aps.org/pra/abstract/10.1103/PhysRevA.93.052105.
Linden, N., Popescu, S. & Wootters, W. Almost every pure state of three qubits is completely determined by its two-particle reduced density matrices. Phys. Rev. Lett. 89, 207901 (2002).
Linden, N. & Wootters, W. The parts determine the whole in a generic pure quantum state. Phys. Rev. Lett. 89, 277906 (2002).
Diósi, L. Three-party pure quantum states are determined by two two-party reduced states. Phys. Rev. A 70, 010302 (2004).
Chen, J., Ji, Z., Ruskai, M. B., Zeng, B. & Zhou, D.-L. Comment on some results of erdahl and the convex structure of reduced density matrices. J. Math. Phys. 53, 072203 (2012).
Chen, J., Ji, Z., Zeng, B. & Zhou, D. From ground states to local hamiltonians. Phys. Rev. A 86, 022339 (2012).
Chen, J. et al. Uniqueness of quantum states compatible with given measurement results. Phys. Rev. A 88, 012109 (2013).
Qi, X.-L. & Ranard, D. Determining a local Hamiltonian from a single eigenstate. Quant. 3, 159 (2019). https://quantum-journal.org/papers/q-2019-07-08-159/.
Hou, S.-Y. et al. Determining system hamiltonian from eigenstate measurements without correlation functions. Preprint at arXiv:1903.06569 (2019).
Qi, B. et al. Quantum state tomography via linear regression estimation. Sci. Rep. 3, 3496 (2013).
Zeng, B., Chen, X., Zhou, D.-L. & Wen, X.-G. Quantum information meets quantum matter–from quantum entanglement to topological phase in many-body systems. Preprint at arXiv:1508.02595 (2015). https://www.springer.com/gp/book/9781493990825.
Kieferová, M. & Wiebe, N. Tomography and generative training with quantum boltzmann machines. Phys. Rev. A 96, 062327 (2017).
Torlai, G. et al. Neural-network quantum state tomography. Nat. Phys. 14, 447 (2018).
Chandra, P. & Singh, Y. Fault tolerance of feedforward artificial neural networks—a framework of study. In Proc. International Joint Conference on Neural Networks, Vol. 1, 489–494 (IEEE, 2003). https://ieeexplore.ieee.org/document/1223395.
Singh, Y. & Chauhan, A.S. Neural networks in data mining. J. Theor. Appl. Inf. 5, 37–42 (2009).
Basheer, I. A. & Hajmeer, M. Artificial neural networks: fundamentals, computing, design, and application. J. Microbiol. Methods 43, 3–31 (2000).
Le Roux, N. & Bengio, Y. Representational power of restricted Boltzmann machines and deep belief networks. Neural Comput. 20, 1631–1649 (2008).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Proc. 3rd International Conference on Learning Representations (ICLR, 2015). https://dblp.uni-trier.de/db/conf/iclr/iclr2015.html.
Reddi, S. J., Kale, S., and Kumar, S. On the convergence of Adam and beyond. Proc. 6th International Conference on Learning Representations (ICLR, 2018).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436 (2015).
Nielsen, M. A. & Chuang, I. Quantum Computation and Quantum Information (Cambridge University Press, 2002).
Fortunato, E. M. et al. Design of strongly modulating pulses to implement precise effective hamiltonians for quantum information processing. J. Chem. Phys. 116, 7599–7606 (2002).
Xin, T. et al. Nuclear magnetic resonance for quantum computing: techniques and recent achievements. Chinese Phys. B 27, 020308 (2018).
Vandersypen, L. M. & Chuang, I. L. NMR techniques for quantum control and computation. Rev. Mod. Phys. 76, 1037 (2005).
Jones, J. A., Vedral, V., Ekert, A. & Castagnoli, G. Geometric quantum computation using nuclear magnetic resonance. Nature 403, 869 (2000).
Xin, T. et al. Nmrcloudq: a quantum cloud experience on a nuclear magnetic resonance quantum computer. Sci. Bull. 63, 17–23 (2018).
Cory, D. G., Fahmy, A. F. & Havel, T. F. Ensemble quantum computing by NMR spectroscopy. Proc. Natl Acad. Sci. USA 94, 1634–1639 (1997).
Fahmy, A. F. & Havel, T. F. Nuclear magnetic resonance spectroscopy: an experimentally accessible paradigm for quantum computing. In Quantum Computation and Quantum Information Theory: Reprint Volume with Introductory Notes for ISI TMR Network School, Physica D: Nonlinear Phenomena, Vol. 120, Issues 1–2, 82–101 (1998). https://doi.org/10.1016/S0167-2789(98)00046-3.
Knill, E., Chuang, I. & Laflamme, R. Effective pure states for bulk quantum computation. Phys. Rev. A 57, 3348 (1998).
Schmidhuber, J. Deep learning in neural networks: an overview. Neural Net. 61, 85–117 (2015).
Chollet, F. et al. Keras. https://keras.io (2015).
Abadi, M. et al. Tensorflow: a system for large-scale machine learning. In Proc. of the 12th USENIX conference on Operating Systems Design and Implementation, Vol. 16, 265–283 (2016). https://dl.acm.org/citation.cfm?id=3026899.
Nair, V. & Hinton, G. E. Rectified linear units improve restricted boltzmann machines. In ICML'10 Proc. of the 27th International Conference on International Conference on Machine Learning, (eds Fürnkranz, J. & Joachims, T.) 807–814 (2010). https://dl.acm.org/citation.cfm?id=3104425.
James, D. F., Kwiat, P. G., Munro, W. J. & White, A. G. On the measurement of qubits. In: Asymptotic Theory of Quantum Statistical Inference: Selected Papers (ed. Hayashi, M. (Japan Science and Technology Agency & University of Tokyo)) 509–538 (World Scientific, 2005).
Hradil, Z. & Řeháček, J. Efficiency of maximum-likelihood reconstruction of quantum states. Fortschr. Phys. 49, 1083–1088 (2001).
Řeháček, J., Hradil, Z., Knill, E. & Lvovsky, A. Diluted maximum-likelihood algorithm for quantum tomography. Phys. Rev. A 75, 042108 (2007).
Paris, M. & Rehacek, J. Quantum State Estimation, Vol. 649 (Springer Science & Business Media, 2004).
Acharya, A., Kypraios, T. and Guţă, M. A comparative study of estimation methods in quantum tomography. J. Phys. A: Math Theor. 52(23), 234001, (2019). https://iopscience.iop.org/article/10.1088/1751-8121/ab1958.
MacBook Pro, Processor: 2.3 GHz Intel Core i5, Memory: 8 GB.
Gershenfeld, N. A. & Chuang, I. L. Bulk spin-resonance quantum computation. Science 275, 350–356 (1997).
Boulant, N., Edmonds, K., Yang, J., Pravia, M. & Cory, D. Experimental demonstration of an entanglement swapping operation and improved control in NMR quantum-information processing. Phys. Rev. A 68, 032305 (2003).
Khaneja, N., Reiss, T., Kehlet, C., Schulte-Herbrüggen, T. & Glaser, S. J. Optimal control of coupled spin dynamics: design of nmr pulse sequences by gradient ascent algorithms. J. Magn. Reson. 172, 296–305 (2005).
Ryan, C., Negrevergne, C., Laforest, M., Knill, E. & Laflamme, R. Liquid-state nuclear magnetic resonance as a testbed for developing quantum control methods. Phys. Rev. A 78, 012328 (2008).
Lu, D. et al. Enhancing quantum control by bootstrapping a quantum processor of 12 qubits. npj Quantum Inf. 3, 45 (2017).
Leskowitz, G. M. & Mueller, L. J. State interrogation in nuclear magnetic resonance quantum-information processing. Phys. Rev. A 69, 052302 (2004).
Lee, J.-S. The quantum state tomography on an NMR system. Phys. Lett. A 305, 349–353 (2002).
Li, J. et al. Optimal design of measurement settings for quantum-state-tomography experiments. Phys. Rev. A 96, 032307 (2017).
Altepeter, J. B., Jeffrey, E. R. & Kwiat, P. G. Photonic state tomography. Adv. Atom. Mol. Opt. Phys. 52, 105–159 (2005).
Song, C. et al. 10-qubit entanglement and parallel logic operations with a superconducting circuit. Phys. Rev. Lett. 119, 180511 (2017).
Krizhevsky, A., Sutskever, I. & Hinton, G.E. Advances in Neural Information Processing Systems 25. In Advances in Neural Information Processing Systems (eds Pereira, F., Burges, C. J. C., Bottou, L. & Weinberger, K. Q.) 1097–1105 (Curran Associates, Inc., 2012). https://dl.acm.org/citation.cfm?id=3065386.
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition, 770–778 (2016).
Flammia, S. T., Gross, D., Liu, Y.-K. & Eisert, J. Quantum tomography via compressed sensing: error bounds, sample complexity and efficient estimators. New J. Phys. 14, 095022 (2012).
Riofrío, C. et al. Experimental quantum compressed sensing for a seven-qubit system. Nat. Commun. 8, 15305 (2017).
Acknowledgements
We thank Yi Shen for helpful discussions. G.L. is grateful to the following funding sources: the National Natural Science Foundation of China (11175094); National Basic Research Program of China (2015CB921002). T.X., D.L., and J.L. are supported by the National Natural Science Foundation of China (Grants nos. 11605153, 11605005, 11875159, U1801661, 11905099, and 11975117), Science, Technology and Innovation Commission of Shenzhen Municipality (Grants nos. ZDSYS20170303165926217 and JCYJ20170412152620376), Guangdong Innovative and Entrepreneurial Research Team Program (Grant no. 2016ZT06D348). T.X. is also supported by Guangdong Basic and Applied Basic Research Foundation (No. 2019A1515011383). N.C. and B.Z. acknowledge the Natural Sciences and Engineering Research Council of Canada (NSERC), Canadian Institute for Advanced Research (CIFAR) and Chinese Ministry of Education (20173080024).
Author information
Authors and Affiliations
Contributions
B.Z. conceived the idea of this paper. S.L. and N.C. wrote and implemented the computer code and simulations. T.X. accomplished the NMR experiments. B.Z. supervised the project. T.X., S.L., and N.C. wrote the manuscript with feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xin, T., Lu, S., Cao, N. et al. Local-measurement-based quantum state tomography via neural networks. npj Quantum Inf 5, 109 (2019). https://doi.org/10.1038/s41534-019-0222-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41534-019-0222-3
This article is cited by
-
Learning quantum systems
Nature Reviews Physics (2023)
-
Quantum process tomography of unitary maps from time-delayed measurements
Quantum Information Processing (2023)
-
Efficient experimental characterization of quantum processes via compressed sensing on an NMR quantum processor
Quantum Information Processing (2022)
-
A machine learning approach to Bayesian parameter estimation
npj Quantum Information (2021)
-
Quantum second-order optimization algorithm for general polynomials
Science China Physics, Mechanics & Astronomy (2021)