Abstract
Recurrent mutations in the SLC12A3 gene responsible for autosomal recessive Gitelman syndrome (GS) are frequently reported, but the exact prevalence is unknown. The rapid detection of recurrent SLC12A3 mutations may help in the early diagnosis of GS. This study was aimed to investigate the prevalence of recurrent SLC12A3 mutations in a Taiwan cohort of GS families and develop a simple and rapid method to detect recurrent SLC12A3 mutations. One hundred and thirty independent Taiwan families with genetically confirmed GS were consecutively enrolled to define recurrent SLC12A3 mutations and determine their prevalence. Using TaqMan probe-based real-time polymerase chain reaction, we designed a mutation detection plate with all recurrent mutations. We validated this mutation detection plate and tested its feasibility in newly diagnosed GS patients. A total of 57 mutations in the SLC12A3 gene were identified and 22 including 2 deep intronic mutations were recurrent mutations consisting of 87.1% (242/278, 18 triple) of all allelic mutations. The recurrent mutation-based TaqMan assays were fully validated with excellent sensitivity and specificity in genetically diagnosed GS patients and healthy subjects. In clinical validation, recurrent mutations were recognized in 92.0% of allelic mutations from 12 GS patients within 4 h and all were confirmed by direct sequencing. Recurrent SLC12A3 mutations are very common in Taiwan GS patients and can be rapidly identified by this recurrent mutation-based SLC12A3 mutation plate.
Similar content being viewed by others
Introduction
Gitelman syndrome (GS) is characterized by persistent renal salt wasting, chronic hypokalemic metabolic alkalosis with renal potassium (K+) wasting, hypomagnesemia with inappropriate renal magnesium (Mg2+) loss, and hypocalciuria. Clinical manifestations of GS vary dramatically from asymptomatic, fatigue, dizziness, numbness, paresthesia, joint pain (pseudogout and chondrocalcinosis), and neuromuscular weakness to paralysis or fatal arrhythmia1,2,3,4. Patients with GS were also found to carry an increased risk of the development of chronic kidney disease (CKD) and type 2 diabetes mellitus5.
GS is inherited as autosomal recessive renal tubulopathy caused by inactivating mutations in the SLC12A3 gene encoding thiazide-sensitive sodium chloride cotransporter (NCC) on the apical membrane of distal convoluted tubule6,7. Given a much higher prevalence of GS carrier (1–4%), GS may be the most common inherited salt-losing tubulopathy with an estimated prevalence of 1–16 in 40,000 (refs. 8,9,10). Although hypocalciuria and hypomagnesemia in the presence of persistent renal salt wasting are landmark findings for GS in the differential diagnosis of chronic hypokalemia, genetic testing is still the gold standard to confirm GS11,12,13.
The SLC12A3 gene on chromosome 16 is composed of 26 exons with >130 kb. To date, approximately 500 different mutations have been identified with a wide distribution along 26 exons and some introns of SLC12A3 (refs. 14,15,16). Direct Sanger sequencing has long been regarded as the gold standard to detect the genetic mutations for GS. However, large genomic rearrangement, large deletion, and deep intronic mutations that were not so rare in GS may not be detected by direct sequencing17. It has been described that recurrent SLC12A3 mutations were highly prevalent among unrelated GS families18,19,20. In a large French cohort of patients with GS, seven recurrent SLC12A3 mutations (p.A313V, p.G394fs (c.1180+1G>T), p.G741R, p.L559P, p.R861C, p.H916fs (c.2883+1G>T), and p.C994Y) contributed to 41.5% of all identified allelic mutations21. We collected a large cohort of GS families5,14 and found that recurrent SLC12A3 mutations occurred frequently with undefined prevalence. The rapid detection of these recurrent NCC mutations, albeit challenging, may provide an early genetic diagnosis of GS.
This study was aimed to define the prevalence of recurrent SLC12A3 mutations from 130 genetically confirmed GS families and design a recurrent mutation-based detection plate with validation using TaqMan probe-based real-time polymerase chain reaction (RT-PCR). In addition, we clinically evaluated the feasibility of the diagnostic plate in newly diagnosed GS patients. The results to be reported indicated that 22 recurrent mutations of the 57 identified SLC12A3 mutations comprised 87.1% of all allelic mutations. Using this simple recurrent mutation-based detection plate in 12 clinically diagnosed GS patients, 92.0% of allelic mutations were rapidly detected.
Results
Identification of SLC12A3 mutations
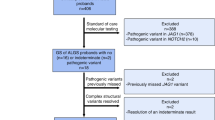
As shown in Fig. 1, 57 different SLC12A3 mutations including 10 novel ones in 130 unrelated GS families were identified. The most common mutation type was a missense mutation (47.5%), followed by frameshift (17.6%), splicing site mutations (17.3%), nonsense mutations (12.6%), and indels (5.0%). The majority of GS patients (112/130, 86.2%) were compound heterozygous. Eighteen GS families (13.8%) had triple mutations with p.T163M and p.R871H being exclusively on one allele at a much higher frequency (66.7%), suggestive of a founder effect (Table 1).
Prevalence of recurrent SLC12A3 mutations
As shown in Fig. 2, 22 recurrent mutations accounted for 87.1% of all allelic mutations (242 of 278 allelic mutations). All GS patients carried at least one recurrent mutation. The most prevalent mutations in order were p.R959fs, p.T60M, c.1670-191C>T, p.S710X, p.T163M, and p.R871H, responsible for 56.5% of the total allelic mutations. Of note, two deep intronic mutations (c.1670-191C>T in intron 13 and c.2548+253C>T in intron 21) that were undetected by conventional genetic tests constituted 13.7% of all allelic mutations.
Characteristics of recurrent SLC12A3 mutations
These 22 recurrent SLC12A3 mutations were located in extracellular loops (7/22), intracellular loops (5/22), C-terminal domain (5/22), N-terminal domain (3/22), and rarely in transmembrane domains (2/22). A mutation (p.T60M) at the phosphorylation site of NCC regulated by STE20/SPS1-related proline/alanine-rich kinase/oxidative stress-responsive kinase-1 (SPAK/OSR1)22 was not only frequent in our cohort but also in Japan, Korea, and western countries18,23,24. Three recurrent mutations with p.D486N, p.R642C, and c.506-1G>A were reported in many different geographic areas18,23,24,25. Twelve recurrent mutations (p.R83Q, p.T163M, p.L215P, p.N442K, p.R642H, p.T649M, p.W844X, p.R871H, p.R959fs, c.602-16G>A, c.1670-191C>T, and c.2548+253C>T) have been reported in Japan18,26,27. Three recurrent mutations (p.T649M, p.W844X, and c.602-16G>A) were detected in western countries25,28,29 Mutation p.H90Y could be found in Korea24.
Validation of recurrent mutation-based mutational detection plate
As shown in Fig. 3, the mutation detection plate employing TaqMan probe-based RT-PCR for 22 recurrent SLC12A3 mutations was created successfully with 100% accuracy. Every assay validated in samples from 130 GS patients and 50 healthy volunteers exhibited fairly high sensitivity (>99%) and specificity (>99%). One healthy volunteer harboring one recurrent mutation also confirmed by Sanger sequencing suggested a higher prevalence of GS carriers in Taiwan’s population.
a The probe is labeled with a 3′ quencher (a dark QSY or a non-fluorescent minor groove binder (MGB) quencher) and a 5′ fluorescent reporter dye (ABY, JUN, FAM, or VIC). During the standard procedure of RT-PCR, the TaqMan polymerase with 5′–3′ nuclease activity will release the fluorescent reporter which will be detected in each cycle. b Example of the output of allelic discrimination software and interpretation of allelic discrimination plot based on the signal measurement of VIC and FAM fluorescent reporter dye. c Interpretation of the results from our p.D486N and p.T60M assays on validation. The target recurrent single-nucleotide variants were inserted into vector pUC57 as positive control for each assay as well as two no-template controls as negative control. As shown with p.T60M, six blue plots represented one positive control and five homozygous samples. The number of green plots and red plots indicated the number of samples carrying one (heterozygous) or no (wild type) allelic mutation p.T60M.
Clinical validation in newly diagnosed GS patients
As shown in Table 2 and Supplementary Fig. 2, 12 newly diagnosed GS patients were included for the detection of recurrent SLC12A3 mutations. Ten (83.3%) of them were identified as biallelic recurrent mutations including one triple mutations. With direct sequencing, the remaining two GS patients with one recurrent mutation were confirmed to carry a novel splicing site mutation c.2285+2T>C (IVS18+2T>C) and p.D62G on the other allele, respectively. Overall, recurrent mutations were recognized in 92.0% (23/25) of allelic mutations.
Discussion
In this study, we identified 57 distinct SLC12A3 mutations with 22 being recurrent in 130 unrelated genetically confirmed Taiwan GS families. The prevalence of these recurrent SLC12A3 mutations was much higher, approximately 87.1% of all allelic mutations. Using TaqMan probe-based RT-PCR, we designed a novel recurrent mutation-based SLC12A3 mutation detection plate that was successfully validated in GS patients and healthy subjects. In the prospective evaluation for 12 newly diagnosed GS patients, recurrent mutations were rapidly identified in 92.0% of all allelic mutations.
GS is one of the most common inherited salt-losing tubulopathies and genetic testing remains the gold standard to confirm the diagnosis11,12,13. With advanced molecular techniques, approximately 500 SLC12A3 mutations have been reported in >1300 GS patients14,15,16. Compound heterozygosity with different mutations on each allele is the most common (up to 70%) with missense mutation being the most frequent type30,31. In our cohort, the prevalence of compound heterozygosity was even higher, approximately 86.2% of GS families. Of note, triple SLC12A3 mutations with one mutation in one allele and two in the other have been increasingly reported28,32. We found that 13.8% of our GS families had triple SLC12A3 mutations and all the mutations were classified as pathogenic according to the ACMG guidelines. Mutations p.T163M and p.R871H exclusively on the same allele in GS patients with triple mutations at a higher frequency suggest linkage disequilibrium secondary to a possible founder effect. The highly prevalent triple SLC12A3 mutations indicate that a complete genetic analysis of family pedigree should still be warranted even though two pathogenic mutations were identified. Besides, this implies that there may be many mutational hotspots on SLC12A3; screening these hotspots will provide a rapid tool in the molecular diagnosis of GS.
Our cohort also showed a significantly higher occurrence of deep mutations within introns 13 and 21, resulting in a pseudo-exon inclusion in GS families33. To date, deep intronic mutations—usually missed by conventional sequencing analyses—have been recognized as causative variants in >75 diseases such as β-thalassemia but were rarely reported in GS patients previously34. The higher prevalence may highlight the importance of searching for deep intronic mutations using cDNA analysis in GS patients if first-line methodologies fail to identify causative mutations.
The meticulous analyses of all reported SLC12A3 mutations to date clearly shows that recurrent mutations in SLC12A3 were relatively common5,24,35,36,37. In Japan, two recurrent mutations (p.R642C and p.L858H) are reportedly responsible for 38.6% of all allelic mutations in GS families37. One European survey enrolling 164 independent families reported that 47 recurrent mutations out of 115 mutations accounted for 77.9% of all allelic mutations23. In this study, 22 recurrent mutations accounted for >87.1% SLC12A3 allelic mutations. In light of the higher prevalence of recurrent mutations in SLC12A3, rapidly identifying them may be very helpful in the early molecular diagnosis of GS.
Routine genetic screening for SLC12A3 mutations is still time-, labor-, and cost-consuming. Several time-saving methods have been developed to detect recurrent mutations in human-inherited diseases or cancer such as neurofibromatosis, epileptic disease, and breast cancer13,38. Allelic-specific TaqMan probe-based RT-PCR is a high-throughput method and has been widely used in virological testing, somatic mutation detection in cancers, and polymorphism screening5,39,40. Its use in identifying recurrent SLC12A3 mutations in GS has not been evaluated. Using this methodology, we successfully created a mutation detection plate and confirmed its fairly high sensitivity and specificity. All mutations of validation samples, including deep intronic mutations, were accurately recognized with easily interpreted results.
A prospective test of this mutation detection plate in 12 newly diagnosed GS patients rapidly identified >90% of allelic mutations. For samples with single or without detectable recurrent mutations, we performed a step-by-step approach and identified two additional non-recurrent SLC12A3 mutations on the other allele in two GS patients. Considering that the majority of recurrent mutations in our plate are recognized frequently in different geographic regions, we suggested that using the mutation detection plate as the primary screen for GS should improve the efficacy of GS diagnosis. Furthermore, this recurrent mutation-based detection approach may also be applied to other genetic diseases with a high prevalence of recurrent mutations such as hemophilia A, cystic fibrosis, and bilateral sensorineural hearing loss41,42.
Compared to other high-throughput platforms, our detection plate does not need to run cases in batches. The cost of our mutation detection plate is around 30 USD/patient with a rapid turnaround time (within 4 h), much cheaper and efficient than either targeted panel sequencing, which has a high cost (around 350 USD/patient) and slower turnaround time (5–7 days), or wholeexome sequencing (WES), which has a higher cost, far longer turnaround time (weeks), and requires dedicated professionals. Furthermore, mutations deep in the introns responsible for about 20% of mutant alleles will be missed in WES.
This study has some limitations. First, rare, novel, and non-recurrent mutations could not be recognized by this recurrent mutation-based detection plate. To expand the application, more mutational spots should be added to enhance the diagnostic efficiency. Second, we did not compare the cost-effectiveness between this mutation detection plate and conventionally used sequencing or other genetic analyses.
In conclusion, recurrent mutations in the SLC12A3 gene are highly prevalent in Taiwan GS patients and can be quickly detected by this accurate recurrent mutation-based detection plate. Early detection of SCL12A3 mutations may enhance the medical care quality in GS. Further investigation and validation is still warranted in other cohorts of GS.
Methods
Study design and participants
The study protocol was approved by the Institutional Review Board (B202005082) at Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan. Prior written informed consent was obtained from all subjects and parental assent for any minors from their family members in our study according to the guidelines approved by the ethics vommittee.
In total, 130 unrelated families of 161 probands with genetically confirmed GS were consecutively enrolled over the past 15 years. There were 103 families with one proband, 23 families with two probands, and four families with three probands. Complete genetic analysis of the probands and family members was performed for cosegregation whenever available and also in all GS families with triple SLC12A3 mutations. The enrolled families were not only identified from our hospital but also referred from other hospitals in different regions of Taiwan, including indigenous and Han populations43,44,45,46. All of them have persistent renal salt wasting, chronic hypokalemic metabolic alkalosis with renal K+ wasting (urine K+/Cr ratio >2 mmol/mmol), renal Mg2+ wasting with or without hypomagnesemia, and hypocalciuria or normocalciuria.
Detection of pathogenic mutations in SLC12A3
Using genomic DNA and/or cDNA from blood leukocytes (National Center for Biotechnology Information No. NM-000339), a series of meticulous genetic analyses of SLC12A3 were performed. Genetically confirmed GS was defined as recognition of inactivating SLC12A3 mutations in both alleles. Sanger sequencing was performed first with primers pairs that were designed for exons and introns based on previous studies6,33,47. For those with no biallelic mutations, denatured high-performance liquid chromatography, direct sequencing, multiplex ligation-dependent probe amplification for large deletions, cDNA analysis via RT-PCR for splicing site and deep intronic mutations, and targeted sequencing panel for all genes relevant in the differential diagnosis of GS were performed in order as previously reported14,33,47.
To confirm the pathogenicity of the identified nucleotide variants, we excluded single-nucleotide variants with a frequency of >1% at first using a publicly available database (HapMap, dbSNP150 (http://www.ncbi.nlm.nih.gov/projects/SNP/), the 1,000 Genomes Project (http://www.1000genomes.org), the Genome Aggregation Database (gnomAD, https://gnomad.broadinstitute.org), and the Exome Aggregation Consortium (ExAC) database). Pathogenicity score then was evaluated via in silico computational methods using SIFT, PolyPhen2, LRT, Mutation Taster, Mutation Accessor, FATHMM, M-CAP, CADD, and GERP38,48,49,50. Furthermore, the pathogenicity of all candidate variants was annotated based on American College of Medical Genetics and Genomics (ACMG) standards and guidelines51.
Definition of recurrent SLC12A3 mutations and prevalence
Recurrent SLC12A3 mutation was defined as the reappearance of the same mutation in two or more unrelated families. Triple mutations were defined as two different mutations in one allele and one mutation in another allele. If two or more siblings from one family carried the same mutations, “one occurrence” of each mutation was calculated to prevent the overestimation of prevalence. Accordingly, the prevalence of recurrent mutations was determined as the total number of recurrent mutations divided by the total number of all allelic mutations in 130 “families” rather than patients.
Development of recurrent mutation-based mutation detection plate
To create a recurrent mutation-based detection plate, SLC12A3 mutations that fulfilled the definition of recurrent mutation were obtained. Primers and TaqMan probes were designed by Primer Express software (Applied Biosystems, USA). Two types of dual-labeled probes were applied for RT-PCR in this study. The QSY probes incorporate a dark 3′ QSY quencher and a 5′ fluorescent reporter dye (ABY or JUN). The minor groove binder (MGB) probes contain a non-fluorescent quencher (NFQ) attached to a 3′ MGB moiety and a fluorescent reporter dye (FAM or VIC) incorporated into the 5′ end. Each mutation detection assay included a pair of primers and two allele-specific probes with different reporter dyes corresponding to mutant and wild-type alleles, respectively. Table 3 shows the primer and probe sequences.
Using a 96-well plate, all TaqMan probe-based RT-PCR experiment was performed in a 10 μL reaction mixture consisting of 5 μL 2× TaqMan master mix buffer (Life Technologies), 300 nM of each primer, 300 nM of probes, and 100 ng DNA template. The preparation of the mixtures was carried out on EzMate, an Automated Liquid Handling & Pipetting System. The TaqMan probe-based RT-PCR was done in QuantStudio 5 Real-Time PCR System (Thermo Fisher) with the following thermal cycle program: 60 °C for 30 s followed by 95 °C for 5 min and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Real-time data were collected during 40 cycles of amplification and were analyzed using the TaqMan Genotyper software v.1.3 (Life Technologies).
Validation of all recurrent mutations
To test the function of each mutation detection assay in this recurrent mutation-based mutation detection plate, a blind study was conducted for each assay. DNA samples from all 130 unrelated GS families were included with concealed genotypes and plated in duplicates on the 96-well plates, using 100 ng DNA of each sample as a template in the above-described RT-PCR assay. The genotypes of each sample were uncovered for comparison at the end of this blind study to evaluate the accuracy. Besides, target recurrent mutations were inserted into vector pUC57 as a positive control for each assay as well as two no-template controls as negative controls. When referring to the sensitivity and specificity, each TaqMan assay was accessed in DNA samples including GS patients with or without specific mutations and healthy subjects.
Clinical validation in newly diagnosed GS patients
Patients newly diagnosed as GS over a year were prospectively enrolled to validate this mutation detection plate with Sanger sequencing confirmation. Vector pUC57 carrying homozygous and heterozygous target recurrent mutations was applied concomitantly as a positive control. DNA samples from three healthy subjects and vector pUC57 with wild-type SLC12A3 were also used as wild-type control as well as one no-template control as a negative control (Supplementary Fig. 1). If the detection plate fails to make a definite diagnosis, further genetic analyses as mentioned above were performed.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Ellison, D. H. & Lin, S. H. Enemy action in the distal convoluted tubule. J. Am. Soc. Nephrol. 30, 1345–1348 (2019).
Li, C. et al. Identification of two novel mutations in SLC12A3 gene in two Chinese pedigrees with Gitelman syndrome and review of literature. Clin. Endocrinol. (Oxf.) 83, 985–993 (2015).
Gladziwa, U. et al. Chronic hypokalaemia of adults: Gitelman’s syndrome is frequent but classical Bartter’s syndrome is rare. Nephrol. Dial. Transplant. 10, 1607–1613 (1995).
Cruz, D. N., Shaer, A. J., Bia, M. J., Lifton, R. P. & Simon, D. B. Gitelman’s syndrome revisited: an evaluation of symptoms and health-related quality of life. Kidney Int. 59, 710–717 (2001).
Tseng, M. H. et al. Genotype, phenotype, and follow-up in Taiwanese patients with salt-losing tubulopathy associated with SLC12A3 mutation. J. Clin. Endocrinol. Metab. 97, E1478–E1482 (2012).
Simon, D. B. et al. Gitelman’s variant of Bartter’s syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat. Genet. 12, 24–30 (1996).
Mastroianni, N. et al. Novel molecular variants of the Na-Cl cotransporter gene are responsible for Gitelman syndrome. Am. J. Hum. Genet. 59, 1019–1026 (1996).
Knoers, N. V. & Levtchenko, E. N. Gitelman syndrome. Orphanet J. Rare Dis. 3, 22 (2008).
Hsu, Y. J. et al. Heterozygous mutations of the sodium chloride cotransporter in Chinese children: prevalence and association with blood pressure. Nephrol. Dial. Transplant. 24, 1170–1175 (2009).
Ji, W. et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat. Genet. 40, 592–599 (2008).
Bettinelli, A. et al. Use of calcium excretion values to distinguish two forms of primary renal tubular hypokalemic alkalosis: Bartter and Gitelman syndromes. J. Pediatr. 120, 38–43 (1992).
Matsunoshita, N. et al. Differential diagnosis of Bartter syndrome, Gitelman syndrome, and pseudo-Bartter/Gitelman syndrome based on clinical characteristics. Genet. Med. 18, 180–188 (2016).
Lin, S. H. & Halperin, M. L. Hypokalemia: a practical approach to diagnosis and its genetic basis. Curr. Med. Chem. 14, 1551–1565 (2007).
Blanchard, A. et al. Gitelman syndrome: consensus and guidance from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 91, 24–33 (2017).
Lin, S. H. et al. Phenotype and genotype analysis in Chinese patients with Gitelman’s syndrome. J. Clin. Endocrinol. Metab. 90, 2500–2507 (2005).
Mastroianni, N. et al. Molecular cloning, expression pattern, and chromosomal localization of the human Na-Cl thiazide-sensitive cotransporter (SLC12A3). Genomics 35, 486–493 (1996).
Badalian-Very, G. Personalized medicine in hematology—a landmark from bench to bed. Comput. Struct. Biotechnol. J. 10, 70–77 (2014).
Takeuchi, Y. et al. Exonic mutations in the SLC12A3 gene cause exon skipping and premature termination in Gitelman syndrome. J. Am. Soc. Nephrol. 26, 271–279 (2015).
Coto, E. et al. A new mutation (intron 9 +1 G>T) in the SLC12A3 gene is linked to Gitelman syndrome in Gypsies. Kidney Int. 65, 25–29 (2004).
Shao, L. et al. Novel SLC12A3 mutations in Chinese patients with Gitelman’s syndrome. Nephron Physiol. 108, 29–36 (2008).
Tavira, B. et al. A labor- and cost-effective non-optical semiconductor (Ion Torrent) next-generation sequencing of the SLC12A3 and CLCNKA/B genes in Gitelman’s syndrome patients. J. Hum. Genet. 59, 376–380 (2014).
Yang, S. S. et al. Phosphorylation regulates NCC stability and transporter activity in vivo. J. Am. Soc. Nephrol. 24, 1587–1597 (2013).
Glaudemans, B. et al. Novel NCC mutants and functional analysis in a new cohort of patients with Gitelman syndrome. Eur. J. Hum. Genet. 20, 263–270 (2012).
Lee, J. W., Lee, J., Heo, N. J., Cheong, H. I. & Han, J. S. Mutations in SLC12A3 and CLCNKB and their correlation with clinical phenotype in patients with Gitelman and Gitelman-like syndrome. J. Korean Med. Sci. 31, 47–54 (2016).
Ashton, E. J. et al. Simultaneous sequencing of 37 genes identified causative mutations in the majority of children with renal tubulopathies. Kidney Int. 93, 961–967 (2018).
Nozu, K. et al. A deep intronic mutation in the SLC12A3 gene leads to Gitelman syndrome. Pediatr. Res. 66, 590–593 (2009).
Monkawa, T., Kurihara, I., Kobayashi, K., Hayashi, M. & Saruta, T. Novel mutations in thiazide-sensitive Na-Cl cotransporter gene of patients with Gitelman’s syndrome. J. Am. Soc. Nephrol. 11, 65–70 (2000).
Vargas-Poussou, R. et al. Spectrum of mutations in Gitelman syndrome. J. Am. Soc. Nephrol. 22, 693–703 (2011).
Blanchard, A. et al. Resistance to insulin in patients with gitelman syndrome and a subtle intermediate phenotype in heterozygous carriers: a cross-sectional study. J. Am. Soc. Nephrol. 30, 1534–1545 (2019).
Qin, L. et al. Identification of five novel variants in the thiazide-sensitive NaCl co-transporter gene in Chinese patients with Gitelman syndrome. Nephrology (Carlton) 14, 52–58 (2009).
Ma, J. et al. Genetic features of Chinese patients with Gitelman syndrome: sixteen novel SLC12A3 mutations identified in a new cohort. Am. J. Nephrol. 44, 113–121 (2016).
Liu, T. et al. Genotype/phenotype analysis in 67 Chinese patients with Gitelman’s syndrome. Am. J. Nephrol. 44, 159–168 (2016).
Lo, Y. F. et al. Recurrent deep intronic mutations in the SLC12A3 gene responsible for Gitelman’s syndrome. Clin. J. Am. Soc. Nephrol. 6, 630–639 (2011).
Vaz-Drago, R., Custódio, N. & Carmo-Fonseca, M. Deep intronic mutations and human disease. Hum. Genet. 136, 1093–1111 (2017).
Wang, F., Shi, C., Cui, Y., Li, C. & Tong, A. Mutation profile and treatment of Gitelman syndrome in Chinese patients. Clin. Exp. Nephrol. 21, 293–299 (2017).
Zeng, Y. et al. Genetic analysis of SLC12A3 gene in Chinese patients with Gitelman syndrome. Med Sci. Monit. 25, 5942–5952 (2019).
Fujimura, J. et al. Clinical and genetic characteristics in patients with Gitelman syndrome. Kidney Int. Rep. 4, 119–125 (2018).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Navarro, E., Serrano-Heras, G., Castaño, M. J. & Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 15, 231–250 (2015).
Wang, J. Y. et al. MGB probe assay for rapid detection of mtDNA11778 mutation in the Chinese LHON patients by real-time PCR. J. Zhejiang Univ. Sci. B 9, 610–615 (2008).
Youssoufian, H., Antonarakis, S. E., Bell, W., Griffin, A. M. & Kazazian, H. H. Jr. Nonsense and missense mutations in hemophilia A: estimate of the relative mutation rate at CG dinucleotides. Am. J. Hum. Genet. 42, 718–725 (1988).
Eiklid, K. et al. Frequency of the delta F508 and exon 11 mutations in Norwegian cystic fibrosis patients. Clin. Genet. 44, 12–14 (1993).
Groeneveld, J. H., Sijpkens, Y. W., Lin, S. H., Davids, M. R. & Halperin, M. L. An approach to the patient with severe hypokalaemia: the potassium quiz. QJM 98, 305–316 (2005).
Lin, S. H., Chiu, J. S., Hsu, C. W. & Chau, T. A simple and rapid approach to hypokalemic paralysis. Am. J. Emerg. Med. 21, 487–491 (2003).
Ng, H. Y. et al. Hypokalemic paralysis due to Gitelman syndrome: a family study. Neurology 67, 1080–1082 (2006).
Chan, C. F., Mu, S. C., Lau, B. H., Chang, C. J. & Lin, S. H. Gitelman’s syndrome: report of one case. Acta Paediatr. Taiwan 49, 31–34 (2008).
Riveira-Munoz, E. et al. Transcriptional and functional analyses of SLC12A3 mutations: new clues for the pathogenesis of Gitelman syndrome. J. Am. Soc. Nephrol. 18, 1271–1283 (2007).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Shihab, H. A. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34, 57–65 (2013).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Acknowledgements
We are indebted to the participating patients and their families for their cooperation and thank the physicians and nephrologists for the contribution of patient material and clinical information. The design of mutation detection plate also won the Taiwan National Innovation award in 2019. This study was supported in part by grants from the Ministry of Science and Technology, Taiwan (MOST 106-2314-B-016-035-MY3), the Research Fund of Tri-Service General Hospital (TSGH-C-107-096, TSGH-C-108-132, TSGH-D-109-123 and TSGH-E-110-201), and Ter-Zer foundation for Educational Achievement.
Author information
Authors and Affiliations
Contributions
S.-H.L. and M.-T.Y. designed the study and drafted the pape; S.-S.Y. and M.-T.Y. carried out experiments; S.-H.L., J.-D.T., C.-J.C. and M.-T.Y. collected and analyzed data; C.-C.S. and M.-H.T. made the figures; S.-H.L., C.-J.C., Y.-J.H. and M.-T.Y. revised the paper; all authors reviewed the results and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Consent for publication: All authors have read and approved this article for publication.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yan, MT., Yang, SS., Tseng, MH. et al. Allele-specific RT-PCR for the rapid detection of recurrent SLC12A3 mutations for Gitelman syndrome. npj Genom. Med. 6, 68 (2021). https://doi.org/10.1038/s41525-021-00230-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-021-00230-8