Abstract
Recurrence and clustering of somatic mutations (hotspots) in cancer genomes may indicate positive selection and involvement in tumorigenesis. MutSpot performs genome-wide inference of mutation hotspots in non-coding and regulatory DNA of cancer genomes. MutSpot performs feature selection across hundreds of epigenetic and sequence features followed by estimation of position- and patient-specific background somatic mutation probabilities. MutSpot is user-friendly, works on a standard workstation, and scales to thousands of cancer genomes.
Similar content being viewed by others
Introduction
Cancer is a genetic disease arising from (driver) mutations that give cancer cells a selective advantage to proliferate and invade. Early cancer genomics studies have mainly focused on the protein-coding regions of the genome. However, even with thousands of cancer exomes sequenced in the past decade, identification of putative driver mutations in the coding regions has still not saturated in many cancer types1,2. Importantly, mutations in the non-coding DNA that constitutes the other 98% of the human genome is even less explored. Tumor whole-genome sequencing is, however, gaining popularity and a recent study of over 2500 tumor whole genomes by the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Network (PCAWG) estimated that up to 25% of all tumors harbor non-coding driver mutations3. There is therefore a pressing need to develop statistical methods that can leverage these large datasets to predict driver mutations in the non-coding DNA.
Current tools designed to identify non-coding drivers are based on mutation recurrence within regulatory elements4,5,6, predicted functional impact of somatic mutations7, or a combination of these approaches8,9. However, existing methods are designed to explore mutations within defined regulatory regions, such as promoters, enhancers or UTRs, therefore ignoring the rest of the non-coding genome. As such, a typical non-coding cancer driver detection method evaluates less than 5% of the 3 million bases sequenced in a WGS experiment for signs of positive selection. Furthermore, by restricting the analysis to annotated regulatory regions, current tools will miss non-coding drivers that create de novo regulatory elements in regions of unannotated DNA. For example, non-coding mutation hotspots upstream of TAL1 and LMO1 in T-cell acute lymphoblastic leukemia lead to the formation of de novo MYB binding sites that drives the overexpression of TAL1 and LMO1 oncogenes10,11. Here, we present MutSpot, an R package that systematically and unbiasedly scans the entire genome for mutation hotspots with statistical evidence of positive selection.
Results
Detection of mutation hotspots in gastric cancer genomes
MutSpot can be used to detect mutation hotspots either genome-wide or in user-defined regions. In the genome-wide discovery mode, MutSpot fits a genomic background model and scans for mutation hotspots across the whole genome. In the regional discovery mode, MutSpot fits a background model specific to the user-defined regions, e.g., promoters, and predicts hotspots in the specified regions only. While the genome-wide mode provides a comprehensive scan of the entire genome, the regional mode can be advantageous when the mutational processes in the regions of interest are very different from the genomic background. To demonstrate the utility of the regional analysis, we ran MutSpot on 168 microsatellite stable gastric cancer whole genomes12 to detect SNV hotspots (1) genome-wide and (2) in regions comprising CTCF binding sites (CBS; 47,453 CBSs analyzed). MutSpot identified 160 mutation hotspots genome-wide (2,533,374,732 nucleotides evaluated) and 12 mutation hotspots in CBSs (1,164,231 nucleotides evaluated) at FDR <0.05. In each analysis, MutSpot outputs a Manhattan plot of the detected hotspots and a barplot of the Z-values (quantifying association with mutation rate) of the selected features in the fitted background model (Fig. 1b, c). CBSs are known to be hypermutated in gastrointestinal cancers, with a distinct mutation spectrum enriched in A > G and A > C substitutions12,13. In the genome-wide background mutation model, CpG dinucleotides, individual tumor mutation burden and local mutation rate are among the top predictors of mutation probability. In contrast, and consistent with the current knowledge, MutSpot identifies AA dinucleotides as the most important predictor of mutation probability in the CBS-specific model. Twenty-three mutation hotspots at CBSs are identified in the genome-wide model. However, only 12 remain significant in the CBS-specific model that corrects for the elevated background mutation rate and unique mutation spectrum at CBSs.
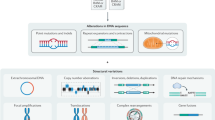
a MutSpot analysis workflow. b, c For each analysis, MutSpot outputs three types of descriptive figures: a Manhattan plot, a feature importance plot of features evaluated by the background mutation model, and lollipop plots of the top hotspots. Figures produced by MutSpot from b a genome-wide analysis and c a CBS-specific analysis of 168 gastric cancer whole genomes. Hotspots with FDR <0.05 are labeled in magenta. d, e Comparison of the number of hotspots detected using MutSpot with the number of hotspots detected using other statistical approaches in d the genome-wide and e CBS-specific analyses.
As there are few validated drivers in the non-coding DNA, we validated the ability of MutSpot to identify known mutation hotspots in the protein-coding regions. MutSpot identified 10 hotspots in four genes using the gastric cancer cohort (Supplementary Fig. 1). All four genes are known drivers of gastric cancer (TP53, CTNNB1, KRAS, and RHOA). There were a total of 38 unique protein-altering mutations in the 10 hotspots, and 37/38 mutations are found to be hotspot mutations by a previously published pan-cancer analysis of protein-coding hotspots2,14.
Performance of MutSpot on other tumor cohorts
The statistical power for detection of hotspots depends on factors such as tumor cohort size and the passenger mutation rate in the specific cancer type1. To further demonstrate the performance of MutSpot, we ran MutSpot on two additional cancer cohorts with different passenger mutation loads. First, we ran MutSpot on 31 paediatric T-cell acute lymphoblastic leukemia (T-ALL) tumors11, using MutSpot default features and lymphocyte-specific epigenetic profiles in the feature selection step (See Supplementary Methods). We identified non-coding hotspots upstream of three known T-ALL oncogenes (LMO1, LMO2, and TAL1; Supplementary Fig. 2), demonstrating that hotspot detection could be useful even in small cancer cohorts. Next, we performed hotspot discovery on 70 melanoma tumors3. The high passenger mutation load in melanoma and the presence of local hypermutation at TF-binding sites (TFBS)15,16 make hotspot detection in melanoma especially challenging. To account for known mutational biases in melanoma, we included melanoma-specific epigenetic and sequence features in addition to the default MutSpot features for feature selection (See Supplementary Methods). MutSpot identified 79 mutation hotspots at 1% FDR, and the top hotspot identified overlaps the two known hotspot mutations in the TERT gene promoter (Supplementary Fig. 3). Melanoma tumors are hypermutated at active TFBSs in gene promoters due to impaired nucleotide excision repair (NER) at these sites15. The default MutSpot model without tissue-specific features predicted 104 mutation hotspots with 47 hotspots overlapping gene proximal TFBSs (Supplementary Fig. 3). Using instead a model also correcting for local hypermutation at active TFBSs in melanoma, only 25 out of the 79 significant hotspots identified were located in active TFBSs in gene promoters. By examining common features of the remaining NER-associated hotspots identified by MutSpot, one could potentially identify additional covariates of the somatic mutation processes acting on these sites. Such features could then be modelled by MutSpot in an iterative manner to further refine the background mutation model to reduce false-positive hotspots.
Comparison to existing methods
We compared MutSpot against other statistical approaches for driver detection adopted by previous studies17,18,19 (Fig. 1d, e, Supplementary Fig. 4). Since none of these approaches are available as standalone software packages for hotspot detection, we implemented four commonly used strategies: (1) Binomial model based on the average genome-wide mutation rate in the cohort, (2) Poisson Binomial model accounting for heterogeneity in genome-wide mutation rates across individual tumors, (3) Poisson Binomial model also correcting for variation in DNA replication timing, and (4) Poisson Binomial correcting for both DNA replication timing and local mutation rate. Expectedly, models that integrated more information about confounding factors predicted fewer candidate hotspots (Fig. 1d, e, Supplementary Fig. 4). Hotspots predicted by only the simpler models are likely false-positives, since their frequency can be explained by genomic covariates of the somatic mutation rate. Overall, this indicates that the larger covariate feature space modelled by MutSpot reduces the number of potential false-positive hits.
A recent study by the PCAWG consortium has examined pan-cancer non-coding drivers using an ensemble of different driver discovery methods20. However, most of these methods were designed to identify positive selection in annotated regulatory regions, and none of the methods work out of the box for genome-wide hotspot detection. To further validate the performance of MutSpot, we adapted three existing methods (OncoDriveFML7, ncdDetect4,21, and ActiveDriverWGS6) for genome-wide hotspot detection by first identifying potential hotspot regions (short windows with four or more mutations) and then used these regions as input for each method (see Supplementary Methods). From the cohort of 168 gastric cancer tumors, 87/90 hotspots identified by MutSpot are also found by at least one other method (Supplementary Fig. 5). Similarly, in the cohort of 70 melanoma samples, 74/79 hotspots identified by MutSpot are found by at least one other method (Supplementary Fig. 5). In summary, MutSpot is currently the only standalone tool available for genome-wide identification of mutation hotspots, and the predictions made by MutSpot are generally concordant with other driver identification methods.
Discussion
MutSpot offers the flexibility to incorporate any genomic or clinical covariate into the background mutation model. This allows users to include tissue-specific epigenetic features for the cancer type of interest, as well as other newly discovered mutational biases into the background mutation model. As our current knowledge of the mutational processes and biases is far from complete, new insights into the processes underlying somatic mutations will further improve the accuracy of hotspot detection.
In conclusion, MutSpot is a user-friendly tool for end-to-end non-coding mutation hotspot identification from cancer genomes. As an increasing number of cancer whole genomes become available, MutSpot can facilitate the discovery of novel drivers in the non-coding genome to further our understanding of tumor biology.
Methods
Input features for modeling background mutation rates
Non-coding hotspots are small, focal regions with high recurrence and clustering of somatic mutations. By default, Mutspot defines a hotspot as a 21 bp region with at least two mutations. To accurately detect mutation hotspots, MutSpot builds a logistic regression model to estimate patient- and position-specific background mutation rates while correcting for known covariates of mutation probability, such as local nucleotide context, replication timing, and epigenomic features12 (Fig. 1a and Table 1). As mutation hotspot detection can be sensitive to recurrent sequencing or variant-calling artifacts, the users are recommended to prefilter the input mutations to remove likely mapping and sequencing errors (see Supplementary Methods). In addition, MutSpot excludes problematic regions, such as poorly mappable regions and immunoglobin loci, from the analysis. Poorly mappable regions are defined as regions with mappability score <1 in the ENCODE 75mers Alignability track in the UCSC genome browser. Separate background mutation models are built for single nucleotide variants (SNVs) and small insertions and deletions (indels), as they arise from different mutational processes. By default, MutSpot automatically computes 763 sequence, epigenetic, and structural features (Table 1). As replication timing profiles and transcription factor (TF) binding profiles are not yet available for many tissue types, MutSpot provides the mean replication profile of 13 ENCODE cell lines, and the aggregate TF-binding profile over all available ENCODE cell lines as default non-tissue-specific features. However, we expect tissue-specific epigenetic profiles to be more predictive of the background mutation rates in individual cancer types22. Therefore, we recommend users to input tissue-specific epigenetic features for feature selection if available. Additional epigenomic features such as DNase I hypersensitive sites (DHSs) and histone modification profiles can be provided by the user in the bigwig or bed format (Table 1). Tissue-specific DHSs and histone modification profiles for a large number of tissues are readily available from the Roadmap Epigenomics Project23.
Feature selection using LASSO regression
The most predictive features of mutation probabilities are selected by a LASSO logistic regression model. MutSpot randomly samples 1 million mutated sites from the input mutation file (or all mutated sites if the total number of mutations is less than 1 million) and an equal number of non-mutated sites as the input for the LASSO logistic regression model. Then, the mutation status of each site is regressed against all candidate sequence or epigenetic features. The regularization parameter is chosen as the value at which the error of the model is within one standard deviation from the minimum, as determined by 10-fold cross-validation. MutSpot performs LASSO regression on 100 bootstrap samples with 50% of the data in each bootstrap, and selects for epigenomic features with more than 75% recurrence frequency and sequence features with more than 90% recurrence frequency. The user can adjust these thresholds to control the number of features included in the final background mutation model. To determine the number of mutations required for optimal performance of feature selection, we repeated LASSO feature selection on the gastric cancer and melanoma cohorts by sampling 50k, 100k, 250k, 500k, 750k, 1 million (default), 1.5 million, and 2 million mutated sites, and an equal number of non-mutated sites in each experiment. Then, we fitted logistic regression models based on features selected in each experiment, and calculated the MacFadden’s pseudo-R2 to estimate the model fit. We found that the MacFadden’s pseudo-R2 levels off at around 200k sampled sites (100k mutated sites) for both cohorts (Supplementary Fig. 6). Overall, we recommend the tumor cohort to have at least 100,000 mutations for optimal performance of feature selection. MutSpot uses the ‘glmnet’ package for LASSO regression and cross-validation.
Sample- and position-specific background mutation model
To account for interpatient heterogeneity, MutSpot corrects for the mutation burden of individual tumors. Additional patient-specific features such as mutation signatures and cancer subtypes can also be integrated into the model. Finally, MutSpot fits a logistic regression model over all positions in the genome to estimate patient- and position- specific background mutation probabilities.
Here, X includes sequence and epigenetic features selected by LASSO regression as well as sample-specific features such as tumor mutation count and clinical features.
Identification of mutation hotspots
To identify mutation hotspots, MutSpot evaluates the mutation recurrence for l-bp regions with at least n mutated samples genome-wide (default l = 21, n = 2). We set the default window size (l) to 21 nucleotides because most TF-binding motifs are shorter than 20 nucleotides, and recurrent mutations that create or abolish a specific TFBS should therefore cluster within 20 bp. The user can set the recurrence parameter n based on the desired minimum recurrence frequency (e.g., set n = 20 for a cohort of 1000 tumors to detect hotspots with at least 2% recurrence). Increasing the recurrence parameter decreases the compute time as fewer regions are evaluated (Supplementary Fig. 7). The p-value of mutation recurrence is computed using a Poisson binomial model that accounts for varying mutation rates across different patient tumors12,18. Multiple hypothesis testing is corrected using the Benjamini Hochberg method. Theoretically, MutSpot evaluates each position in the genome and its 20 bp flank for mutation recurrence, although in practice only regions with at least n mutated samples are evaluated. Non-evaluated nucleotides with fewer than two mutated samples in its 20 bp flanks are assigned P = 1, and P-values are corrected for multiple testing across all nucleotides in the masked non-coding genome (2,533,374,732 nucleotides). In the region-specific mode, the number of hypotheses is the number of nucleotides in the masked regions of interest.
As it can be computationally expensive to fit genome-wide models with multiple covariates, sparse matrices were implemented to minimize memory usage and a multi-threading option is available to reduce the compute time. MutSpot takes less than 3 hours on a 4-core machine for genome-wide hotspot discovery in 200 tumors, and it can be scaled up to process thousands of tumors on a standard workstation (Supplementary Fig. 8).
Preprint
A previous version of this manuscript was published as a preprint24.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Gastric cancer mutation data are available as Supplementary Data 3 of Guo et al12. T-ALL somatic mutations were obtained from Hu et al11. Melanoma somatic mutations are available for download at https://xenabrowser.net/. Roadmap Epigenomics data are available for download at http://www.roadmapepigenomics.org/data/. ENCODE data are available for download at ftp://ftp.ensembl.org/pub/release-85/regulation/homo_sapiens/.
Code availability
MutSpot is implemented as an R package and is available at https://github.com/skandlab/MutSpot/. All R code used to generate the figures and statistics of the paper is included in Supplementary Data.
References
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Chang, M. T. et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016).
Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Juul, M. et al. Non-coding cancer driver candidates identified with a sample- and position-specific model of the somatic mutation rate. elife. https://doi.org/10.7554/eLife.21778 (2017).
Lochovsky, L., Zhang, J., Fu, Y., Khurana, E. & Gerstein, M. LARVA: an integrative framework for large-scale analysis of recurrent variants in noncoding annotations. Nucleic Acids Res. 43, 8123–8134 (2015).
Zhu, H. et al. Candidate cancer driver mutations in distal regulatory elements and long-range chromatin interaction networks. Mol. Cell. https://doi.org/10.1016/j.molcel.2019.12.027 (2020).
Mularoni, L., Sabarinathan, R., Deu-Pons, J., Gonzalez-Perez, A. & Lopez-Bigas, N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 17, 128 (2016).
Dhingra, P. et al. Identification of novel prostate cancer drivers using RegNetDriver: a framework for integration of genetic and epigenetic alterations with tissue-specific regulatory network. Genome Biol. 18, 141 (2017).
Hornshoj, H. et al. Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. npj Genom. Med. 3, 1 (2018).
Mansour, M. R. et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346, 1373–1377 (2014).
Hu, S. et al. Whole-genome noncoding sequence analysis in T-cell acute lymphoblastic leukemia identifies oncogene enhancer mutations. Blood 129, 3264–3268 (2017).
Guo, Y. A. et al. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun. 9, 1520 (2018).
Katainen, R. et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat. Genet 47, 818–821 (2015).
Chang, M. T. et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 8, 174–183 (2018).
Sabarinathan, R., Mularoni, L., Deu-Pons, J., Gonzalez-Perez, A. & Lopez-Bigas, N. Nucleotide excision repair is impaired by binding of transcription factors to DNA. Nature 532, 264–267 (2016).
Fredriksson, N. J. et al. Recurrent promoter mutations in melanoma are defined by an extended context-specific mutational signature. PLoS Genet. 13, e1006773 (2017).
Weinhold, N., Jacobsen, A., Schultz, N., Sander, C. & Lee, W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165 (2014).
Melton, C., Reuter, J. A., Spacek, D. V. & Snyder, M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 47, 710–716 (2015).
Guilhamon, P. & Lupien, M. SMuRF: a novel tool to identify regulatory elements enriched for somatic point mutations. BMC Bioinforma. 19, 454 (2018).
Rheinbay, E. et al. Analyses of non-coding somatic drivers in 2658 cancer whole genomes. Nature 578, 102–111 (2020).
Juul, M. et al. ncdDetect2: improved models of the site-specific mutation rate in cancer and driver detection with robust significance evaluation. Bioinformatics 35, 189–199 (2019).
Polak, P. et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518, 360–364 (2015).
Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Guo, Y. A., Chang, M. M. & Skanderup, A. J. MutSpot: detection of non-coding mutation hotspots in cancer genomes. Prepint at https://www.biorxiv.org/content/10.1101/740944v1 (2019).
Aggarwala, V. & Voight, B. F. An expanded sequence context model broadly explains variability in polymorphism levels across the human genome. Nat. Genet. 48, 349–355 (2016).
Zerbino, D. R., Wilder, S. P., Johnson, N., Juettemann, T. & Flicek, P. R. The ensembl regulatory build. Genome Biol. 16, 56 (2015).
Hansen, R. S. et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl Acad. Sci. USA 107, 139–144 (2010).
Buisson, R. et al. Passenger hotspot mutations in cancer driven by APOBEC3A and mesoscale genomic features. Science. https://doi.org/10.1126/science.aaw2872 (2019).
Acknowledgements
This work was supported by Singapore National Medical Research Council grant OFIRG15nov072. We would like to thank Probhonjon Baruah, Danliang Ho, Weitai Huang, Zhong Wee Poh, and Julie Solacroup for discussion during the development and testing of MutSpot.
Author information
Authors and Affiliations
Contributions
A.J.S. and Y.A.G. designed the study. Y.A.G. and M.M.C. wrote the R package and analyzed the data. Y.A.G. and A.J.S. interpreted the data and wrote the manuscript, with contributions from all authors. Y.A.G. and M.M.C. contributed equally to the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, Y.A., Chang, M.M. & Skanderup, A.J. MutSpot: detection of non-coding mutation hotspots in cancer genomes. npj Genom. Med. 5, 26 (2020). https://doi.org/10.1038/s41525-020-0133-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41525-020-0133-4
This article is cited by
-
Multi-omic profiling of peritoneal metastases in gastric cancer identifies molecular subtypes and therapeutic vulnerabilities
Nature Cancer (2021)
-
Non-coding driver mutations in human cancer
Nature Reviews Cancer (2021)