Abstract
We investigated the association between TP53 mutation and 21-gene recurrence score (RS) in ER-positive/HER2-negative breast cancer (BC) using data from 141 patients who underwent TP53 sequencing and Oncotype DX® tests. We detected TP53 mutations in 18 (12.8%) patients. Most patients with TP53 mutation had a high 21-gene RS (≥26). The average 21-gene RS was higher in TP53 mutant tumors. Multivariate analysis showed that mutated TP53 is an independent factor for a high 21-gene RS. Mutated TP53 remained closely associated with high 21-gene RS in patients with low pathological risk (n = 103). In the ER+/PR+/HER2-negative subset (n = 356) of The Cancer Genome Atlas, the non-luminal A intrinsic subtype was more prevalent in the group with mutant TP53. mRNA levels of p53-regulated senescence gatekeeper and cell cycle-related genes were increased in BC with mutated TP53. Mutational analysis of TP53 helped identify endocrine-resistant tumors.
Similar content being viewed by others
Introduction
The tumor suppressor protein p53 can induce cell cycle arrest, apoptosis, senescence, or ferroptosis in response to stress signals1,2, and it prevents the accumulation of cancer-causing mutations that lead to the development of malignant tumors3. Approximately 30% of the breast tumors harbor a TP53 mutation4, and the frequency, spectrum, and timing of these mutations vary with the molecular subtype of the disease5. Although the prevalence of TP53 mutations is lower in luminal than in basal-like tumors6, TP53 mutation is the second most common mutation in the luminal type7.
TP53 mutation is a poor prognostic factor in hormone receptor (HR)-positive luminal tumors8,9,10. Moreover, 11 (38%) of the 29 aromatase inhibitor-resistant, estrogen receptor (ER)-positive (+) tumors had somatic mutations in genes involved in the TP53 pathway, including TP53, ATR, APAF1, or THBS17, indicating an association between TP53 mutations and endocrine resistance. Somatic TP53 mutations are more prevalent in patients with primary than in patients with secondary endocrine-resistant or -responsive ER+ metastatic breast cancer (BC)11.
The Oncotype DX® 21-gene recurrence score (RS) is a validated multigene signature for predicting outcomes and guiding chemotherapy in ER+/HER2− BC12,13,14. This score has also been evaluated in cohorts given neoadjuvant endocrine therapy or chemotherapy. Patients with a high 21-gene RS respond poorly to neoadjuvant endocrine therapy15,16,17, implying an association between RS and endocrine resistance. The pathological complete response rates in neoadjuvant chemotherapy cohorts are higher when tumors have a high 21-gene RS18,19. Our translational study revealed that tumors with high 21-gene RS are more chemo-sensitive, based on chemoresponse assays in vitro20. These clinical and translation studies suggest an association between high 21-gene RS and endocrine resistance in ER+ BC.
A correlation between the TP53 mutation and 21-gene RS in ER+ BC has not yet been identified. We, therefore, investigated the association between the TP53 mutation and the 21-gene RS in patients with ER+/HER2− BC and evaluated the molecular characteristics of ER+/HER2− BC with the TP53 mutation using The Cancer Genome Atlas (TCGA) database.
Results
Characteristics of patients with TP53 mutations
We identified TP53 gene mutations in 18 (12.8%) of 141 patients with 21-gene RS and a sequenced TP53 gene and studied their types and locations. Nonsense and missense TP53 mutations were the most prevalent types in 9 (50%) and 4 (22.2%) of 18 patients, respectively (Fig. 1). The most frequent locations of these mutations were exons 5 and 8 in 5, (27.8%), and (4 (22.2%) of the 18 patients (Fig. 1). We also detected mutations in introns of the TP53 gene in 2 (11.1%) patients.
We investigated the clinicopathological features according to TP53 mutational status (Table 1). The median age was higher in the group with mutated, than wild-type TP53 (57.5 vs. 47.0 y, p = 0.091; Table 1). Mutated TP53 was significantly associated with menopausal status (p = 0.005), T stage (p = 0.021), histological grade (HG) (p = 0.002), lymphovascular invasion (LVI) (p = 0.018), and Ki-67 levels (p ≤ 0.001). Mutations of TP53 were more frequent among postmenopausal patients with a higher histological grade, Ki-67 > 20%, larger tumors, and LVI. Chemotherapy was more frequently administered to patients with, than without the TP53 mutation (p < 0.001; Table 1).
Mutant TP53 and Oncotype Dx® 21-gene RS
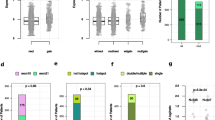
The average 21-gene RSs were 30 in and 16.41 in groups with mutant and wild-type TP53, respectively (Fig. 2a). The 21-gene RS was more likely to be ≥ 26 in tumors with—than without—a TP53 mutation (p = 0.021; Fig. 2b).
a Mean Oncotype DX® RS according to mutated TP53 compared using Student’s t-tests (p < 0.001). Error bars correspond to standard error of the mean. b Distribution of Oncotype DX® risk group compared based on TP53 mutation using Fisher’s exact tests (p = 0.021). Oncotype DX® RS ≥ 26 indicates high risk.
We identified factors associated with a high 21-gene RS using binary logistic regression analysis. Univariate analyses showed that T stage, progesterone receptor (PR), HG, Ki-67 levels, and TP53 mutation were significant (Table 2), and multivariate analysis revealed TP53 mutation, T stage, and PR as independent variables that were associated with a high 21-gene RS (Table 2). The odds ratio (OR) of the TP53 mutation was 21.632 (95% confidence interval [CI], 5.877–79.627). Figure 3 shows the differences between the 21-gene RS and other pathological variables based on the TP53 mutation.
Median age and 21-gene recurrence scores based on TP53 mutation compared using Mann–Whitney U tests. Distribution of Oncotype DX risk group compared according to mutated TP53 using χ2 tests. Histological grade, progesterone receptor status, and Ki-67 expression were compared using Fisher’s exact tests.
TP53 mutation in subset with good pathologic features
We investigated the relationship between mutated TP53 and the 21-gene RS in the subset with good pathological features. The 21-gene RS can determine patients who are eligible for chemotherapy among those with favorable pathological parameters who are nevertheless at high risk of relapse. We compared the clinical characteristics of 103 patients who were PR-positive and had low HG (I/II) and low (≤20%) Ki-67 expression based on TP53 mutations (Supplementary Table 1). Median age, menopausal status, and T stage were altered by the TP53 mutation, whereas other pathological parameters were not.
Tumors in this subset with the TP53-mutant had a higher mean 21-gene RS and an elevated high 21-gene RS rate (Supplementary Fig. 1). Our multivariate analysis selected the TP53 mutation as the sole significant factor for a high 21-gene RS (Supplementary Table 2).
Distant recurrence-free survival according to mutated TP53
We analyzed distant recurrence-free survival (DRFS) to determine the prognostic value of mutated TP53. Six patients had distant recurrence at a median follow-up of 51 (6–98) months. Figure 4a shows that the DRFS was significantly lower in the group with, than without a TP53 mutation (p = 0.046). In addition, the results were consistent; the DRFS significantly differed according to the TP53 mutation in a subset with good pathological features (Fig. 4b; n = 103, p < 0.001) and in a group that had received only endocrine therapy (Fig. 4c; n = 104, p = 0.046). However, the DRFS did not significantly differ according to 21-gene RS ≥ 26 vs. < 26 (Supplementary Fig. 2).
Mutant TP53 in HR+/HER2− stage I/II BC tumors from TCGA
Because mutant TP53 was associated with a high 21-gene RS in the subset with good pathological features, we investigated the molecular characteristics of mutated TP53 in ER+/PR+/HER2−, stage I/II BC tumors using TCGA database.
We identified 356 patients with ER+/PR+/HER2−, stage I/II BC tumors in TCGA database (Fig. 5 and Supplementary Table 3). We compared those with defined PAM50 subtypes. The prevalence of the luminal A subtype was higher in the group with wild-type TP53, whereas that of luminal B, HER2-enriched, and basal-like tumors, was higher in the group with mutant TP53 (p < 0.001). We then compared MDM2/4 amplification between the two groups, because MDM2/4 is a p53-specific E3 ubiquitin ligase that limits the p53 growth-suppressive function in unstressed cells21,22. The frequency of MDM2/4 amplification was comparable between the groups.
We compared the mRNA levels of p53-regulated, senescence gatekeeper, and cell cycle genes. The senescence gatekeeper genes, CCNB1, E2F1, FOXM1, and MYBL2, and the cell cycle-associated genes, HMMR, DLGAP5, BUB1, BIRC5, and AURKB, were upregulated in the group with mutant TP53 (Fig. 5).
Discussion
To the best of our knowledge, this is the first study to investigate the relationship between mutant TP53 and the 21-gene RS. We associated mutant TP53 with a high 21-gene RS (≥ 26) in ER+/HER2− BC. The average 21-gene RS and the frequency of high categorical 21-gene RS were significantly higher in the group with mutant, than wild-type TP53. Therefore, having a TP53 mutation was an independent factor for a high 21-gene RS.
The association between the two markers was reproducible in pathologically low-risk subgroups such as PR+/histological grade (HG) 1/2, and low Ki-67 subgroups. The 21-gene RS helps to identify patients with ER+ BC who are at genomic high and low risk13,23. Mutant TP53 or a high 21-gene RS can both function as biomarkers for endocrine resistance7,11,15,16,17,24,25,26. Our findings showed that TP53 mutational analysis could help to detect endocrine-resistant tumors in patients with good pathological features.
We compared the molecular characteristics of early ER+/PR+/HER2- BC tumors based on TP53 mutational status using TCGA database. Because the 21-gene RS has been principally evaluated in early ER+ BC with limited nodal involvement13,27, we included stage I/II BC. Among the defined PAM50 subtypes, we found that TP53 mutant tumors had non-luminal A (low endocrine sensitivity, chemosensitive) rather than luminal A (endocrine-sensitive) subtypes28,29,30,31. This indicated that TP53 mutant tumors are likely to be endocrine resistant.
We also quantified the mRNA levels of p53-regulated genes associated with the cell cycle or transcription. Upregulated TP53 induces or represses many genes involved in cell cycle regulation, DNA repair, apoptosis, and senescence2,32. Our finding of upregulated gene clusters in the mutant TP53 group indicated that TP53-mutant, ER+ tumors are highly proliferative and aggressive. The upregulated CCNB1 and MYBL2 genes are among those listed for the 21-gene RS assay33. This, to some extent, supports an association between the two biomarkers. Further molecular studies are required to validate an association between genes in the 21-gene RS and those in the p53 signaling pathway.
The amplification rates of MDM2/4 were comparable in tumors with wild-type and mutant TP53. One study found a slightly higher frequency of MDM2 amplification in tumors with mutant, than wild-type TP5334. However, that study considered BC regardless of subtypes, whereas we investigated only ER+/PR+/HER2− BC. Considering ongoing efforts to target MDM2/X in breast cancer35,36, an association between MDM2/4 amplification and TP53 mutational status should be further explored.
A major limitation of the present study was the small sample size in the institutional database and the absence of information on 21-gene RS in TCGA. Due to the small patient cohort and a short follow-up, a prognostic assessment using mutant TP53 combined with 21-gene RS was not obtainable. Whether adding information about mutant TP53 to the 21-gene RS could improve the prognostic ability should be evaluated in a larger patient cohort. Biological and clinical characteristics were not sorted based on the location and type of TP53 mutation37. Nevertheless, our findings underscored the fact that TP53 is associated with a high 21-gene RS in ER+/HER2− BC, and TP53 mutational analysis detected endocrine-resistant tumors.
We concluded that the clinical and molecular evidence points toward an association between mutated TP53 and a high 21-gene RS in terms of endocrine resistance. By detecting TP53 mutation, endocrine-resistant tumors could be identified even in patients who had good pathological features.
Methods
Patients
This study was approved by the Institutional Review Board of Gangnam Severance Hospital (3-2021-0296) and adhered to the clinical practice guidelines of the Declaration of Helsinki (2013 amendment). In all patients, informed consent form for research of human-derived material was obtained. We retrospectively evaluated 572 patients who were surgically treated for ER+/HER2− primary invasive BC who had undergone Oncotype DX® tests at Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea, between August 2011 and March 2020. Among the 572 patients, we included 141 whose TP53 gene had been sequenced. Clinicopathological data were extracted from electronic medical records and included age, menopausal status, ER and PR status, tumor size, nodal status, HG, nuclear grade (NG), LVI, Ki-67, Oncotype DX® 21-gene RS, TP53 mutation status, and some TP53 mutation characteristics. We excluded patients diagnosed with recurrent or metachronous BC. Tumors were staged according to the 7th edition of the American Joint Committee on Cancer. Tissue sections were histologically assessed using the Elston-Ellis modification of the Scarff−Bloom−Richardson grade. Adjuvant systemic therapy and/or radiotherapy were administered according to standard guidelines based on the age of the patients, tumor characteristics, and axillary lymph node status. Endocrine therapy was administered to all patients.
We used specific ER antibodies (1:100 clone 6F11; Novocastra, Newcastle upon Tyne, UK) and the progesterone receptor (PR; clone 16; Novocastra) for immunohistochemistry (IHC)38. The IHC results for ER and PR were stratified based on modified Allred scoring, in which scores of 0–1, 2–4, 5–6, and 7–8 represented negative, weak, moderate, and strong expression, respectively39. Groups with strong and moderate ER expression were considered ER-high and groups with weak expression were considered ER-low. Scores from 2 to 8 (weak to strong) were all considered PR positive.
TP53 gene sequencing
We determined genomic variants using a next-generation sequencing (NGS) panel of 143 genes, including TP53. Among 141 patients, we analyzed 69 (48.9%) using polymerase chain reaction (PCR)-denaturing high-performance liquid chromatography (DHPLC), and 72 (51.1%) using targeted NGS (Supplementary Fig. 3). We analyzed TP53 mutations using PCR-DHPLC and direct sequencing until December 2016 and targeted NGS thereafter.
Mutations in exons 5–9 of the TP53 gene were analyzed by PCR as described40,41, using primers designed to amplify the exons and flanking introns of the TP53 gene40. Amplification proceeded using Accu-Power™ Premix (Bioneer, Daejeon, Korea) under the following cycling conditions: 94 °C for 4 min, 50 cycles of 94 °C for 1 min, 60 °C for 30 s, and 72 °C for 30 s, and then 72 °C for 15 min. Purified PCR products obtained using QIAquick® Gel Extraction kits (Qiagen, Düsseldorf, Germany) were sequenced using BigDye™ Terminator Cycle Sequencing Ready Reaction kits (Applied Biosystems, Foster City, CA, USA) under the following cycling conditions: 96 °C for 5 min, 24 cycles at 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min, then 72 °C for 5 min. The sequences were analyzed using an ABI 3500Dx system (Thermo Fisher Scientific Inc.). Forward and reverse strands were sequenced to confirm mutations.
We applied targeted NGS using an Oncomine™ comprehensive panel and an Ion Torrent™ S5 XL system (Thermo Fisher Scientific Inc.). We extracted DNA from fresh tissues and determined its yield and quality using Torrent v. 5.2 and Ion Reporter™ v. 5.2 (both from Thermo Fisher Scientific Inc.). Genomic variants were analyzed using an NGS panel of 143 genes, including TP53. Supplementary Figure 4 shows other genomic alterations in 72 patients assessed using targeted NGS.
Oncotype Dx® assays
We calculated 21-gene RSs using Oncotype Dx® assays23,33. RSs are derived from the reference-normalized expression of 16 genes associated with cancer (Ki-67, STK15, Survivin, [BIRC5], CCNB1 [cyclin B1], MYBL2, GRB7, HER2, ER, PGR, BCL2, SCUBE2, MMP11 [stromelysin 3], CTSL2 [cathepsin L2], GSTM1, CD68, and BAG1) and five reference genes, β-actin (ACTB), GAPDH, GUS, RPLPO, and TFRC, and then calculated on a scale of 0–100. Quantitative single gene scores were determined by reverse transcriptase-PCR. The expression of each gene was measured in triplicate and normalized to the reference genes. Oncotype Dx® assays of RNA extracted from formalin-fixed paraffin-embedded tissues proceeded at Genomic Health Inc. (Redwood City, CA, USA). We defined scores >26 as high 21-gene RSs according to the TAILORX trial13.
TCGA data
We identified HR+/HER2− tumors in TCGA database using immunohistochemistry (IHC) for ER, PR, and HER2 and RNA-seq data as described42,43 and normalized, log2-transformed, and median-centered expression. Information about TP53 mutations and genomic variants was downloaded from the cBioPortal44. We identified PAM50-defined intrinsic subtypes as luminal A, luminal B, HER2-enriched, basal-like, and normal-like43,45, and compared the subtype distribution and MDM2/4 amplification between the groups with mutant and wild-type TP53. We also quantified the expression of p53-regulated senescence gatekeeper and cell cycle genes35.
Statistical analysis
Continuous variables were compared using Mann–Whitney U tests and Student’s t-tests. The normal distribution of continuous variables was assessed using Kolmogorov–Smirnov tests. Nominal variables were compared using χ2 or Fisher exact tests. Predictive factors for high 21-gene RS (≥26)13 were identified by multivariate binary logistic regression analysis of all variables. Variables with p < 0.05 were included in the multivariate model, and the final model was realized using backward stepwise (Wald) selection. We analyzed survival using DRFS defined as the interval from curative surgery to the first distant recurrence or last censored. Survival was determined using Kaplan–Meier plots, and two groups were compared using log-rank tests. All data were statistically analyzed using SPSS version 25.0 (IBM Corp, Armonk, NY, USA) and R software (https://www.r-projet.org; version 3.6.1). Values with p < 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The TCGA data analyzed within this study are described in the following data record: https://doi.org/10.1038/nature11412 (2012), https://doi.org/10.1016/j.cell.2015.09.033 (2015) and https://doi.org/10.1126/scisignal.2004088 (2013)42,43,44. The somatic mutations and gene amplification data are included as Supplementary Table 4 and presented in Supplementary Fig 4. For more data access requests, please contact the corresponding author, Dr. Sung Gwe Ahn.
References
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008).
Sullivan, K. D., Galbraith, M. D., Andrysik, Z. & Espinosa, J. M. Mechanisms of transcriptional regulation by p53. Cell Death Differ. 25, 133–143 (2018).
Levine, A. J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 20, 471–480 (2020).
Børresen-Dale, A. L. TP53 and breast cancer. Hum. Mutat. 21, 292–300 (2003).
Dumay, A. et al. Distinct tumor protein p53 mutants in breast cancer subgroups. Int. J. Cancer 132, 1227–1231 (2013).
Bertheau, P. et al. p53 in breast cancer subtypes and new insights into response to chemotherapy. Breast 22, S27–S29 (2013).
Ellis, M. J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012).
Meric-Bernstam, F. et al. Survival outcomes by TP53 mutation status in metastatic breast cancer. JCO Precis. Oncol. https://doi.org/10.1200/po.17.00245 (2018).
Lopez, G. et al. Molecular insights into the classification of luminal breast cancers: The genomic heterogeneity of progesterone-negative tumors. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20030510 (2019).
Griffith, O. L. et al. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat. Commun. 9, 3476 (2018).
Hagio, K. et al. Impact of clinical targeted sequencing on endocrine responsiveness in estrogen receptor-positive, HER2-negative metastatic breast cancer. Sci. Rep. 11, 8109 (2021).
Sparano, J. A. et al. Prospective validation of a 21-gene expression assay in breast cancer. N. Engl. J. Med. 373, 2005–2014 (2015).
Sparano, J. A. et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med. 379, 111–121 (2018).
Sparano, J. A. et al. Clinical and genomic risk to guide the use of adjuvant therapy for breast cancer. N. Engl. J. Med. 380, 2395–2405 (2019).
Iwata, H. et al. Validation of the 21-gene test as a predictor of clinical response to neoadjuvant hormonal therapy for ER+, HER2-negative breast cancer: The TransNEOS study. Breast Cancer Res. Treat. 173, 123–133 (2019).
Ueno, T. et al. Evaluating the 21-gene assay Recurrence Score® as a predictor of clinical response to 24 weeks of neoadjuvant exemestane in estrogen receptor-positive breast cancer. Int. J. Clin. Oncol. 19, 607–613 (2014).
Akashi-Tanaka, S. et al. 21-Gene expression profile assay on core needle biopsies predicts responses to neoadjuvant endocrine therapy in breast cancer patients. Breast 18, 171–174 (2009).
Gianni, L. et al. Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J. Clin. Oncol. 23, 7265–7277 (2005).
Yardley, D. A. et al. A phase II trial of ixabepilone and cyclophosphamide as neoadjuvant therapy for patients with HER2-negative breast cancer: Correlation of pathologic complete response with the 21-gene recurrence score. Breast Cancer Res. Treat. 154, 299–308 (2015).
Ahn, S. G. et al. Chemosensitivity to doxorubicin of ER-positive/HER2-negative breast cancers with high 21-gene recurrence score: A study based on in vitro chemoresponse assay. PLoS One 12, e0187679 (2017).
Moll, U. M. & Petrenko, O. The MDM2-p53 interaction. Mol. Cancer Res. 1, 1001–1008 (2003).
Badciong, J. C. & Haas, A. L. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J. Biol. Chem. 277, 49668–49675 (2002).
Paik, S. et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 24, 3726–3734 (2006).
Berns, E. M. et al. Complete sequencing of TP53 predicts poor response to systemic therapy of advanced breast cancer. Cancer Res. 60, 2155–2162 (2000).
Bai, H. et al. Prognostic value of the TP53 mutation location in metastatic breast cancer as detected by next-generation sequencing. Cancer Manag. Res. 13, 3303–3316 (2021).
Ungerleider, N. A. et al. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 20, 115 (2018).
Kalinsky, K. et al. Abstract GS3-00: First results from a phase III randomized clinical trial of standard adjuvant endocrine therapy (ET)+/− chemotherapy (CT) in patients (pts) with 1–3 positive nodes, hormone receptor-positive (HR+) and HER2-negative (HER2-) breast cancer (BC) with recurrence score (RS) < 25: SWOG S1007 (RxPonder). Cancer Res. 81, GS3-00-GS03-00 (2021).
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009).
Gnant, M. et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: Using the PAM50 risk of recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann. Oncol. 25, 339–345 (2014).
Coates, A. S. et al. Tailoring therapies–improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 26, 1533–1546 (2015).
Ades, F. et al. Luminal B breast cancer: Molecular characterization, clinical management, and future perspectives. J. Clin. Oncol. 32, 2794–2803 (2014).
Braithwaite, A. W., Royds, J. A. & Jackson, P. The p53 story: Layers of complexity. Carcinogenesis 26, 1161–1169 (2005).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
Silwal-Pandit, L. et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin. Cancer Res. 20, 3569–3580 (2014).
Portman, N. et al. MDM2 inhibition in combination with endocrine therapy and CDK4/6 inhibition for the treatment of ER-positive breast cancer. Breast Cancer Res. 22, 87 (2020).
Fan, Y. et al. Dual-target MDM2/MDMX inhibitor increases the sensitization of doxorubicin and inhibits migration and invasion abilities of triple-negative breast cancer cells through activation of TAB1/TAK1/p38 MAPK pathway. Cancer Biol. Ther. 20, 617–632 (2019).
Olivier, M. et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin. Cancer Res. 12, 1157–1167 (2006).
Ahn, S. G. et al. Comparison of standardized uptake value of 18F-FDG-PET-CT with 21-gene recurrence score in estrogen receptor-positive, HER2-negative breast cancer. PLoS One 12, e0175048 (2017).
Harvey, J. M., Clark, G. M., Osborne, C. K. & Allred, D. C. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J. Clin. Oncol. 17, 1474–1481 (1999).
Kim, H. W. et al. Patterns and biologic features of p53 mutation types in Korean breast cancer patients. J. Breast Cancer 17, 1–7 (2014).
Ahn, S. G. et al. Low PR in ER(+)/HER2(−) breast cancer: High rates of TP53 mutation and high SUV. Endocr. Relat. Cancer 26, 177–185 (2019).
The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Ciriello, G. et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163, 506–519 (2015).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013).
Garcia-Recio, S. et al. FGFR4 regulates tumor subtype differentiation in luminal breast cancer and metastatic disease. J. Clin. Invest. 130, 4871–4887 (2020).
Acknowledgements
This work was supported by funds from the Basic Science Research Program through the NRF (NRF-2019R1C1C1002830), Republic of Korea. We thank Editage (www.editage.co.kr) for English language editing.
Author information
Authors and Affiliations
Contributions
Study concept and design: J.H.J., J.J., K.L., Y.K., and S.G.A. Data acquisition, analysis, and interpretation: J.H.J., J.J., S.J.B., C.C., K.L., Y.K., J.H.K., and S.G.A. Statistical analysis: J.H.J., S.J.B., C.C., K.L., Y.K., and S.G.A. Drafting the manuscript: J.H.J., J.J., S.J.B., K.L., Y.K., and S.G.A. First author: J.H.J. Corresponding author: S.G.A. All authors reviewed and approved the final draft of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ji, J.H., Bae, S.J., Kim, K. et al. Association between TP53 mutation and high 21-gene recurrence score in estrogen receptor-positive/HER2-negative breast cancer. npj Breast Cancer 8, 19 (2022). https://doi.org/10.1038/s41523-022-00384-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41523-022-00384-3
This article is cited by
-
Extensive review on breast cancer its etiology, progression, prognostic markers, and treatment
Medical Oncology (2023)