Abstract
The neural functions of adropin, a secreted peptide highly expressed in the brain, have not been investigated. In humans, adropin is highly expressed in astrocytes and peaks during critical postnatal periods of brain development. Gene enrichment analysis of transcripts correlating with adropin expression suggests processes relevant to aging-related neurodegenerative diseases that vary with age and dementia state, possibly indicating survivor bias. In people aged <40 y and ‘old-old’ (>75 y) diagnosed with dementia, adropin correlates positively with genes involved in mitochondrial processes. In the ‘old-old’ without dementia adropin expression correlates positively with morphogenesis and synapse function. Potent neurotrophic responses in primary cultured neurons are consistent with adropin supporting the development and function of neural networks. Adropin expression in the ‘old-old’ also correlates positively with protein markers of tau-related neuropathologies and inflammation, particularly in those without dementia. How variation in brain adropin expression affects neurological aging was investigated using old (18-month) C57BL/6J mice. In mice adropin is expressed in neurons, oligodendrocyte progenitor cells, oligodendrocytes, and microglia and shows correlative relationships with groups of genes involved in neurodegeneration and cellular metabolism. Increasing adropin expression using transgenesis improved spatial learning and memory, novel object recognition, resilience to exposure to new environments, and reduced mRNA markers of inflammation in old mice. Treatment with synthetic adropin peptide also reversed age-related declines in cognitive functions and affected expression of genes involved in morphogenesis and cellular metabolism. Collectively, these results establish a link between adropin expression and neural energy metabolism and indicate a potential therapy against neurological aging.
Similar content being viewed by others
Introduction
Aging associates with declining cognitive performance and capacity for tracking the “what/where/when” in our daily activities (episodic memory)1,2,3. Mild Cognitive Impairment (MCI) describes a condition in which impairments in organizing and remembering daily activities are perceptible to individuals but are not severe enough to cause loss of independence4,5. Increased risk for dementia and observations of dementia-related changes in the brain suggests MCI is an early stage in the spectrum of life-threatening dementias, of which Late-Onset Alzheimer’s Disease (LOAD) is the most common6,7,8,9. Demographic changes resulting in an aging population this century will increase the prevalence of MCI and LOAD10. Identifying new treatments targeting aging-related changes in brain structure that underly dementia is thus considered an important goal.
Short open reading frames encoding small polypeptides are a potentially rich source of new drug targets11,12. Adropin1-76 is encoded by the Energy Homeostasis Associated (ENHO) gene, and was identified by Genentech’s Secreted Protein Discovery Initiative13. Modelling in silico suggests a signal sequence (adropin1-33) targets the secretory pathway14,15,16. The predicted secreted domain (adropin34-76) is sufficient for biological activity in cultured cells and rodent models14,17,18,19,20,21,22,23,24,25,26. Reports of adropin immunoreactivity in the circulation of humans, nonhuman primates (NHP) and rodents are consistent with a secreted peptide with endocrine functions14,27,28. However, the exact sequence and any posttranslational modifications of the mature peptide are not known.
Experiments using mouse models suggest adropin regulates metabolic processes in the periphery14,19,26,29,30. However, several observations suggest adropin is a neuropeptide. Expression of the ENHO transcript is orders of magnitude higher in the brain relative to non-neural tissues14,28,31,32,33,34. The two candidates for cell-surface adropin receptors are also highly expressed in the CNS, and each appears to or is known to regulate neural development. The orphan G protein coupled receptor GPR19 is obligatory for adropin activity21,24,25 and is also highly expressed in the CNS relative to other tissues35,36. High GPR19 expression in the embryonic brain suggests a role in development37. However, evidence for coupling of adropin with GPR19 is controversial38. Adropin also interacts with NB-3/Contactin6 (CNTN6) in protein-protein yeast two hybrid assays31. CNTN6 is a brain-specific non-canonical membrane-tethered Notch1 ligand that belongs to the immunoglobulin cell adhesion molecule superfamily (IgCAMs). Contactins function as cell-adhesion proteins in the developing nervous system and regulate axonal guidance, development of neurites, synapse formation, synaptic plasticity, and neural regeneration39.
Recent studies suggest adropin may regulate processes in the nervous system that could affect aging. Aging-related declines of plasma adropin concentrations and protein expression in the brain of Sprague Dawley rats correlate with the expression of protein markers of oxidative stress40. Plasma concentrations in humans also decline with aging, particularly in males41. In NHP tissues, expression of the adropin transcript correlates with genes associated with aging-related neurodegeneration28. Here, we investigate the relationships between adropin expression and aging-related cognitive decline. Results from the analysis of open access transcriptome from humans and intervention experiments using mouse models indicate that adropin is a potential lead for developing new treatments againsts cognitive decline commonly observed with advanced aging.
Results
ENHO expression in human tissue samples
We first compared ENHO expression between human tissues using The Atlas of the Developing Human Brain (www.brainspan.org) and Genotype-Tissue Expression (GTEx) projects. ENHO is highly expressed in all brain structures relative to other tissues (Fig. 1A) and is moderately (10%) higher in some areas of the male brain when compared to females (Fig. S1, S2A). ENHO expression is highest in mature astrocytes, with lower expression in fetal astrocytes, neurons, oligodendrocytes, and endothelial cells (Fig. 1B)42. ENHO expression appears to peak in the first decade of life (Fig. 1C) and is then constant until the 8th decade of life (Fig. 1C and in GTEx data not shown).
A ENHO transcript expression (TPM) in human tissues ranked high to low reading left to right (GTEx). B ENHO expression in isolated cell populations (brain.org). C ENHO expression by age group. This analysis used data from 41 individuals (mean ± SD for age, 15.8 ± 11.2 y, range 4 mo–40 y; 22 males, 14.3 ± 11.7 y, 4 mo–37 y; 19 females, 17.5 ± 10.6 y, 1–40 y) downloaded from The Atlas of the Developing Human Brain28,30. Multiple samples from different brain regions were obtained from individuals; mean expression for samples from individuals was used for the analysis. Data were pooled into 10-year bins (birth-9 y, 10–19 y, 20–29 y, and >30 y). *p < 0.05 vs. all other groups combined. D–E Expression of APOE (D), Clusterin/ApoJ (E), GCK3B (F), and AKT3 (G) related to ENHO (524 samples from 41 individuals, data from The Atlas of the Developing Human Brain28,30).
ENHO expression correlates with energy metabolism and LOAD risk genes in the human brain
A correlation matrix was used to identify gene networks co-regulated with the ENHO transcript using data downloaded from The Atlas of the Developing Human Brain28,30 (Fig. 2A). These data are from 41 individuals (mean ± SD age, 15.8 ± 11.2 y, range 4 mo–40 y); 22 were male (14.3 ± 11.7 y, 4 mo–37 y) and 19 were female (17.5 ± 10.6 y, 1–40 y). Samples from multiple brain structures were obtained for each individual. For the correlation analysis the entire dataset (524 samples) was used. Coefficients (r) comparing ENHO and all other genes expressed were ranked high to low by r. Using r > 0.7 as an arbitrary cut-off to define genes exhibiting similar expression profiles identified 838 genes meeting the selection criteria.
A Correlation matrix heat map showing the relationships between z-scores for ENHO expression and the brain transcriptome. The matrix is representative and was derived from The Atlas of the Developing Human Brain. Genes with a mean expression level (RPKM) of >10 were selected; each pixel is a bin comprised of the averages of ten samples, with ten genes in each sample. B Correlation coefficients (r) between expression of ENHO and the top 1% of genes. ENHO expression shows a tighter correlation, indicated by higher r, in brain samples isolated from the old-old who diet with a dementia diagnosis (‘D’) compared to the old-old without a dementia diagnosis (‘ND’). C Venn diagram showing overlap in the top 500 genes correlating with ENHO expression (ranked by correlation coefficient) for the ND group, D group and people aged < 40 years (< 40 y). D Comparison of biological processes correlating with ENHO expression in the ND group, D group and people aged < 40 years. A score > 0.1 indicates a significant correlation between groups; the D and < 40 y groups appear to be more closely related. E, F GO:Biological Processes correlating with ENHO expression in ND group (D) or D groups (E). ‘#’refers to the number of hits (genes); processes with an adjusted p value of < 0.01 are shown. Biological processes are color coded (orange for neural development, green for metabolism, yellow for signaling).
Experiments using mouse and organ explants suggests that adropin regulates cellular energy metabolism17,18,26. Gene enrichment analysis suggests ENHO expression correlates positively with processes related to reduction-oxidation (redox) reactions and mitochondrial activity. Top ranked GO:molecular functions included “oxidoreductase activity” (q value FDR, 2.11 × 10−18; 86 gene hits/783 genes in genome), “electron transfer activity” (2.37 × 10−10, 28/151), “oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor” (8.08 × 10−7, 15/61), and “NAD(P)H dehydrogenase (quinone) activity” (2.69 × 10−5, 12/49). Top GO:biological processes included “small molecule catabolic process” (2.22 × 10−11, 52/452), and “generation of precursor metabolites and energy” (6.99 × 10−11, 58/571). Top ranked GO:cellular components were mitochondrial: “mitochondrion” (8.04 × 10−23, 153/1897) “mitochondrial inner membrane” (4.62 × 10−18, 68/556), “organelle inner membrane” (5.44 × 10−17, 70/617) and “mitochondrial envelope” (8.98 × 10−17, 83/842).
A candidate gene approach identified strong positive correlations between ENHO and APOE or clusterin (CLU) (Fig. 1D, E). APOE variants (e2, e3, e4) are the strongest genetic risk factors identified for LOAD, with the e4 allele increasing risk and e2 allele conferring protection43,44. Clusterin (CLU, also known as ApoJ) is an extracellular chaperone linked to Aβ clearance and toxicity45. APOE and CLU are highly expressed by astrocytes and, as predicted based on expression data showing high ENHO expression in astrocytes (Fig. 1B), the gene set correlating with ENHO was highly enriched for astrocytic and glial markers (for e.g., ToppCell Atlas “Visual_Cortex-Non-neuronal-Astrocyte|Visual_Cortex/Region, Lineage, Class, Subclass and Cluster” q value FDR, 2.65 × 10–98; 85 hits out of 195 identified genes).
Strong negative correlations were also observed between ENHO and either GSK3B (Fig. 1F) or AKT3 (Fig. 1G), which is the predominant AKT signaling isoform upstream of GSK3B expressed in the brain46. Tau phosphorylation by glycogen synthase kinase 3 (GSK3) contributes to development of neurofibrillary lesions observed in LOAD47.
ENHO relationships with other genes in the ‘old-old’ are specific for dementia status
Whether advanced aging and dementia affects ENHO expression was investigated using data from The Aging, Dementia and TBI Study (http://aging.brain-map.org/). This study involved 107 people selected from the Adult Changes in Thought (ACT) cohort48, a population-based, prospective analysis of neurological aging49,50,51,52. Selected participants were mostly male (63 males, 44 females), varied in years of education and dementia status and age (77–102 y at time of death, median 90 y); 52 were diagnosed with dementia (D) by the time of death (30 with AD; 12 with dementia of multiple etiologies; four with vascular dementia). Postmortem samples were collected from brain regions known to exhibit neurodegeneration and pathology resulting from LOAD and Lewy body disease: frontal white matter (FWM), hippocampus (HIP), parietal cortex, and temporal cortex (TCx).
We first compared ENHO expression between brain structures and between individuals based on dementia state and APOE allele. ENHO expression is not affected by dementia status in humans (Fig. S2B). While these data suggest neurodegenerative diseases do not associate with large differences in ENHO expression, a trend (p < 0.1) was noted for higher ENHO expression in TCx and FWM samples from participants with >1 APOEe4 allele (Fig. S2C). Correlations between brain structures in the expression of gene networks suggests regulation by systemic factors48. We observed correlations in ENHO expression between brain structures (Fig. S2D-K), suggesting that common transcriptional and/or post-transcriptional regulatory elements affect expression throughout the nervous system.
The correlation approach was used to identify gene networks including ENHO to groups divided by dementia diagnosis at the time of specimen collection (Dementia or No Dementia, hereafter referred to as D and ND). The rationale for separating data into two groups is based on relationships between neurodegenerative disease state, RNA quality and agonal conditions that can result from ischemia; these conditions can complicate gene expression data analysis in postmortem brain samples48,53,54.
Aging adversely affect histone structure and transcription factor function55,56. In the old-old group, very few genes met the previously used selection criteria (r > 0.7) (Fig. 2B), consistent with declining fidelity in gene regulation. We therefore used the top 1% of genes (500 out of ~50,200) correlating with the ENHO transcript. Surprisingly, correlation coefficients between ENHO and genes ranked in the top 1% are significantly higher in people with D compared to ND (Fig. 2B). Significant overlap is observed in genes correlating positively with ENHO in the D and ND groups, with 40% (198/500) genes within each population found in both clusters (Fig. 2C). Comparing the top 500 genes correlating with ENHO in the The Atlas of the Developing Human Brain (people aged < 40 y) suggests that a closer relationship exists with the D group relative ND group (Fig. 2C). Out of 500 genes, 92 (18%) were common to the <40 y and D groups while only 52 (10%) were common to the <40 y and ND groups. A comparison of the biological processes correlating with ENHO also indicated a closer relationship between the D and <40 y groups (Fig. 2D).
Using stringent selection criterion (adj. p < 0.05) for gene enrichment analysis indicated differences in biological processes correlating with ENHO in the D and ND groups. In the ND group, neural development dominated the list of biological processes (Fig. 2E). GO:Cellular Component for the D group also related to neurons such as “somatodendritic compartment”. In contrast, metabolic processes dominated in the D group (Fig. 2F). It was also interesting to note processes related to vascular function and development (for e.g., “circulatory system development”, “tube morphogenesis” and “blood vessel development”) are observed in the D group. This is relevant given data from animal and clinical studies showing a positive relationship between adropin expression and vascular function40,57,58,59. GO:Cellular Component for the D group appeared to be predominantly mitochondrial (“mitochondrion”, “mitochondrial membrane”) and cellular processes related to the endoplasmic reticulum such as “organelle development” and “Golgi apparatus”).
To avoid bias associated with selecting small groups of genes, an unbiased screen applied multiple enrichment tools to the entire dataset of genes. As before, data from the D and ND groups were separated for the analysis (http://pharmacology.slu.edu/results/andrew/TBI_Dementia/). Genome-wide gene set enrichment revealed biological pathways either enriched or depleted in a manner correlated with ENHO expression. Four independent studies were performed to include (i) all 5529 biological pathways, designated “All pathways”, or a subset of these pathways belonging to (ii) REACTOME, designated “REACTOME pathways”, (iii) KEGG, designated “KEGG pathways”, or (iv) the remaining pathways, designated “non-REACTOME-KEGG pathways”. Pathways positively correlated with ENHO expression were shown by positive NESs (normalized enrichment scores). Pathways negatively correlated with ENHO expression shown by negative NESs.
The results from this analysis also identified strong correlations between adropin expression and energy metabolism. Specifically, ENHO expression positively associates with catabolic processes such as “KEGG_OXIDATIVE_PHOSPHORYLATION”, “KEGG_FATTY_ACID_METABOLISM” and “REACTOME_RESPIRATORY_ELECTRON_TRANSPORT”. In contrast, negative associations included anabolic pathways such as “KEGG_STEROID_BIOSYNTHESIS”, “KEGG_RIBOSOME”, “KEGG_DNA_REPLICATION”, and “REACTOME_CHOLESTEROL_BIOSYNTHESIS”. Interestingly, high ENHO expression was also associated with low expression of inflammatory pathways such as “REACTOME_INFLAMMASOMES” and “REACTOME_THE_NLRP3_INFLAMMASOME”. ENHO expression in the human brain also correlated with common neurodegenerative disorders (Parkinson’s disease, Huntington’s disease, and Alzheimer’s disease). This was shown by the significant positive or negative enrichment of “KEGG_PARKINSONS_DISEASE”, “KEGG_HUNTINGTONS_DISEASE” and “KEGG_ALZHEIMERS_DISEASE”, or the related pathways in ENHO-expressing or ENHO-depleted patient samples.
The correlations between ENHO expression and neurotrophic processes suggests functions related to brain development, an observation consistent with deficits in neural development reported in adropin knockout mice31. ENHO expression in ‘old-old’ humans appears to associate with increased expression of genes involved in mitochondrial and neurotrophic processes, suggesting that higher expression may benefit patients with advanced age. Indeed, enrichment of neurodegenerative pathways in genes correlating with ENHO expression appears stronger in D patients compared to ND patients. Enrichment of these pathways also appears stronger in female compared to male patients, suggesting sex differences in the robustness of gene networks that include adropin in the ‘old-old’ brain.
ENHO expression correlates with aging-related neuropathology
The Aging, Dementia and TBI Study includes protein markers of dementia-related pathologies and inflammation by immunohistochemistry (IHC) on fresh frozen and formalin fixed paraffin embedded (FFPE) postmortem tissue samples48. In a simple bivariate correlation analysis, levels of ENHO transcript correlated positively with tau pathologies (AT8, Aβ plaques, AB40, pTau and pTau/Tau ratio) in the ND group (Fig. 3A–D), but less so in the D group (Fig. 3E–H). These relationships were further investigated by dividing participants into two groups ranked by ENHO expression above or below the median (Fig. 3I–O). The groups with high ENHO expression appear to exhibit more severe Tau-related pathologies, particularly in participants with no dementia. However, no relationships are observed with inflammatory markers (for example with TNFa, MCP1 and IL1b, Fig. 4). There was however no correlation between ENHO expression (averaged for all four structures) and semi-quantitative scores of AD pathology (CERAD, BRAAK, NIA REAGAN) (data not shown).
Data are shown as scatterplots for samples from study participants with no dementia (A–D) or with dementia (E–H). The box-and-whisker plots shown in (I-O) show the averaged data for D and ND samples split into subgroups (Lo, Hi). ‘Lo’ indicates data from samples with ENHO values below the median (76 samples for D, 90 samples for ND), while ‘Hi’ indicates data from samples with ENHO values above the median (76 samples for D, 90 samples for ND). The markers shown reading top to bottom are AT8 (tau phosphorylation of Ser202 and Thr205), TAU (total phosphorylated tau, and ratio of phosphorylated to non-phosphorylated tau), Aβ, and AB40 (Ab40). Correlation coefficients are shown where associations are significant (p < 0.05). For AT8 and Ab staining, data are shown from fresh frozen (ffpe) or paraffin-embedded sections. The data were subdivided into two groups (below or above the median) after being ranked by ENHO expression. The median is shown as the red dashed line in the scatterplots shown in (A–H). Ranking occurred for all participants (All), or for subgroups separated by dementia status (ND, no dementia; D, dementia). Significantly different from low dementia status, **p < 0.01; *p < 0.05.
Neutrophic properties of adropin in primary cultured hippocampal neurons
To investigate whether variation in adropin expression has causal effects on neurological aging we used primary culture cells and mouse models. Adropin34-76 exhibits dose-dependent trophic properties in primary cultured mouse hippocampal neurons (Fig. 5A–G). Phase contrast images taken on day 2 (Fig. 5A) and day 14 (Fig. 5B) and neurite tracing analyses using ImageJ neurite tracing methods60,61,62 revealed adropin34-76 significantly promoted neuritogenesis (p < 0.001) (Fig. 5C). The number and thickness of branches in neurons were also significantly increased (p < 0.05) (Fig. 5D, E).
A–B Primary cultured hippocampal neurons were cultured in the absence (control) and presence of 1,10 and 100 nM adropin. Phase contrast images were taken at day 2 (early network; A) and day 14 (mature network; B). C Neurons cultured in the presence of 10 and 100 nM adropin for 2 days exhibited longer neurites than neurons without adropin added. The average total neurite outgrowth per phase contrast image of day 2 control neurons was 3125.7 ± 550.5 µm, 1 nM adropin was 4668.3 ± 365.9 µm, 10 nM adroprin was 6599.4 ± 520.7 µm, and 100 nM adropin was 6201.9 ± 791.7 µm (p < 0.01 for control vs. 1 nM adropin and control vs. 100 nM adropin; n = 8 images analyzed per condition). D Addition of 10 nM and 100 nM adropin also significantly increased the number of neuritic branches compared to control neurons. The average number of branches per control image was 72.8 ± 14.0, for 1 nM adropin was 107.5 ± 9.6, for 10 nM adropin was 155.1 ± 19.7, and for 100 nM adropin was 143.8 ± 24.5 (p < 0.05 for control vs. 1 nM adropin and control vs. 100 nM adropin; n = 8 images analyzed per condition). E Neurite thickness was analyzed on neurons cultured for 14 days and showed that 10 nM adropin significantly increased neurite thickness. The average thickness of the five thickest neurites per control image was 5.1 ± 0.3 µm, per 1 nM adropin image was 5.6 ± 0.3 µm, per 10 nM adropin image was 6.8 ± 0.3 µm, and per 100 nM adropin image was 6.0 ± 0.3 µm (p < 0.001 for control vs. 1 nM adropin; n = 5 thickest neurites measured and averaged in eight images per condition to equal 40 neurites analyzed per condition). F Hippocampal neurons cultured on a MEA chip. G Higher magnification image showing electrode and neurons. H–J Sample traces of neuronal spiking activity before and after exposure to adropin (100 nM) using the multi channel recording system. Sample traces demonstrate that adropin increased the spontaneous firing activity in neurons either previously quiescent (H, I) or active (J). K Neuronal spiking activity was quantified as “spikes per minute” and normalized to baseline spikes per minute as described in the Methods. Addition of 100 nM adropin significantly increased the spiking frequency of the neurons. Normalized spikes per minute after 100 nM adropin was 3.2 ± 0.1 spikes per minute (p < 0.0001; n = 31 electrodes). Data are presented as mean ± S.E.M. * indicates p < 0.5, ** indicates p < 0.01, *** indicates p < 0.001, and **** indicated p < 0.0001.
To determine if adropin regulates neuronal excitability, hippocampal neurons were cultured on the non-invasive MEA for recording of neuronal network activity (Fig. 5F, G). After 9 days of culture, adropin (100 nM) was added and neuronal activity recorded before (baseline) and after treatment. The frequency of neuronal activity was significantly increased in hippocampal neurons regardless of baseline activity. Hippocampal neurons that were quiescent at baseline (Fig. 5H–I) began firing action potentials after treatment. In neurons showing spontaneous activity, application of adropin significantly increased firing frequency (spikes/min; Fig. 5J). Statistical data for the recordings (Fig. 5K) demonstrated a neuroexcitatory effect of adropin on hippocampal neurons.
Adropin expression in the C57BL/6J (B6) mouse brain
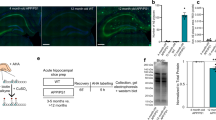
In Sprague Dawley rats adropin protein levels in plasma and brain extracts are lower in 18-month old relative to 4-month old animals40. Here we report that Ehno expression in the peaks between the 1st and 5th week of life in the cortex, HIP and striatum (Fig. 6A)63. Adropin protein levels determined by western blot using brain samples are also lower in 18-month-old B6 mice compared to 4-month-old animals (Fig. 6B, C).
A Expression profiling in mouse brain structures by age show a peak at postnatal day 32 (P32). B, C Adropin immunoreactivity in brain lysates of AdrTG mice compared to wild type (WT) littermates at 4 and 18 months of age. WT mice show a decline in adropin protein levels at 18 months that is not observed in AdrTG. HSP90 was used as a loading control. D Profiling of Enho expression in isolated cells from the mouse brain. E The top panel is a schematic of the approach used to generate an ENHO-Cre reporter mouse. The lower panels (F–Q) are representative photomicrographs from hippocampal sections (F–K) and high magnification photomicrographs of vessels in the cortex (L–Q). Scale bars (10 μM) are shown in the bottom left in (L–Q).
Open access data suggest less cell-type specificity in ENHO expression in cells isolated from the mouse brain compared to humans (Fig. 6D)64. High expression is observed in astrocytes, neurons and oligodendrocyte precursor cells. Lower expression is observed in newly formed and myelinating oligodendrocytes, and in microglial/macrophages. Low expression is also observed in endothelial cells (Fig. 6D)64. Cell-specific patterns of Enho expression were further investigated using B6 mice with IRES-Cre inserted into the Enho locus crossed with the CAG-lox-STOP-lox-tdTomato reporter strain (Fig. 6E). Cre-induced expression of tdTomato was observed throughout the mouse brain (Fig. 6F–Q). In the mouse HIP expression was also observed in neurons, but surprisingly not in astrocytes (Fig. 6F–Q). Expression was also observed in cortical neurons and endothelial (CD31+ve) cells (Fig. 6L–Q), in agreement with experiments using mouse and human endothelial cell lines20,59,65.
Genomic data from the aged human brain indicates inverse associations between the expression of adropin and genes involved in mitochondrial, neurotrophic, and inflammatory processes. These relationships were investigated in the mouse brain using data from open access transcriptome databases and mouse experiments performed at the Saint Louis University.
We first analyzed RNA seq data from experiments involving two lines of transgenic mice used in the investigation of Alzheimer’s disease (GSE125957)66. These transgenic lines overexpress either a human mutant of tau (P301L) or amyloid precursor protein (K670N/M671L and V717F). Enho expression was not identified as a differentially expressed transcript between genotypes66. However, gene enrichment analysis using an r > 0.7 as the selection criteria indicates that Enho expression correlates with pathways also observed in humans (Fig. 7A). GO:Molecular Function terms overlapped between the two populations and were related to protein translation (for e.g., “Ribosome”, “Cytosolic Ribosomal Proteins”) and mitochondria (“oxidative phosphorylation”, “Electron Transport Chain”). Enho expression also correlated with pathways associated with neurodegenerative diseases (“Parkinson’s disease”, “Alzheimer’s disease” and “Huntington’s disease). It is also noteworthy that the strength of the correlations, particularly those related to mitochondrial function, were stronger in transgenic strains expressing mutant proteins causing LOAD in humans (Fig. 7A).
A Gene enrichment analysis of genes correlating with the ENHO transcript in wild type (n = 63) or transgenic mouse models used in Alzheimer’s disease (n = 58) (GSE125957). Pathways correlated with ENHO expression were similar between genotypes and are related to mitochondrial processes and synthesis of large macromolecules. B Heat map showing genes increased or reduced following adropin treatment of 18-month-old male B6 mice with saline or adropin34-76 for 1 month (n = 8/group). These mice were selected from the animals used in the behavioral studies shown in Fig. 8. C Results from gene enrichment analysis showing biological processes responding to adropin34-76 treatment.
The positive correlations between Enho and either Apoe or Clu observed in humans appears to be retained in the mouse brain (Fig. S3), although only Clu meets the selection criteria (r > 0.7). However, genes involved in insulin signaling did not show the same relationship (Fig. S3). Moreover, astrocyte markers were not enriched in the genes correlating with Enho in the mouse brain. This result is consistent with a less cell-specificity in the mouse nervous system (Fig. 6D).
Effects of adropin treatment on the aged mouse brain transcriptome
Transgenic mice over expressing adropin1-76 (AdrTG) under the control of a human b-actin promoter14,17,30,41 were used investigate whether variation of adropin expression affects genes in pathways correlating with ENHO in the human brain. The mouse model used for the experiment (male B6 mice aged 18 months) was selected based on the development of cognitive impairment at or around this age which corresponds to a human age of 56–69 y67.
Increased adropin protein levels are observed in brain lysates from AdrTG mice compared to WT controls, particularly at 18-months of age (Fig. 6B). The results from analysing gene expression using qRT-PCR suggested that adropin protects against neuroinflammation (Fig. 8A–C). Specifically, the expression of genes involved in inflammation were lower in the HIP (Fig. 8A) and the cortex (Fig. 8B) of AdrTG compared to age-matched controls. Measurement of serum cytokines indicated a significant difference (p < 0.05) in IL-6 concentrations, with 60% lower levels in AdrTG mice (Fig. 8C). However, circulating levels of TNFA (Fig. 8C) and other cytokines were not significantly different (Fig. S4). We also assessed for tau phosphorylation and markers of oxidative stress. However, the expression of these markers was highly variable and mostly below the limits of detection (data not shown), likely explained by the mild model used for the study (18-month-old male B6 mice). The expression of genes involved in other biological processes were also not significantly different in AdrTG (Fig. 7A, B).
A, B Reduced expression of genes involved in inflammation in the hippocampus (A) and cortex (B) of male 18-month-old AdrTG mice (n = 13) compared to age-matched controls (n = 12). Expression data (mean, SD) reported as a ratio of WT controls for each gene against 3 ‘housekeeping’ reference genes (Hprt1, 36b4, Ppib). C Serum levels of IL-6 but not TNFA are lower in AdrTG compared to controls. Significance is indicated by *(p < 0.05), **(p < 0.01), or ***(p < 0.001). D–E Improved performance in tests of spatial learning and memory (aversive T-maze) (D) or novel object recognition (E) of 18-month-old AdrTG (n = 9) or B6 mice treated with adropin34-76 for 2 weeks (n = 14) compared to the respective age-matched control group (WT littermates for AdrTG, n = 10; mice treated with diluent for the adropin34-76 treatment group; n = 15). Significance is indicated by *(p < 0.05), **(p < 0.01), or ***(p < 0.001).
The putative secreted domain (adropin34-76) is sufficient for biological activity when administered by ip. injection to mice, inducing changes in mitochondrial fuel selection of skeletal muscle14,17,18,19,30. To investigate whether adropin34-76 administered also regulates the brain transcriptome, RNA seq was used in 18-month-old male B6 mice treated with synthetic peptide at a dose previously shown to be effective. Mice were split into two weight matched groups (Fig. S5A), acclimated to handling and then treated with adropin34-76 (90 nmol/kg/d) suspended in 0.9% saline plus 0.1% BSA over 4 weeks. Controls were treated with diluent; injections were given daily at 0900 h. Applying a cut-off using a fold-change > 1.5 and a p value of <0.05 identified a small number of responsive genes (84 upregulated, 172 downregulated) (Fig. 7B). Gene enrichment analysis indicated that adropin34-76 treatment primarily changed the expression of genes involved in tissue morphogenesis (e.g., “pattern specification process”, “embryo development”, “animal morphogenesis”) (Fig. 7C). Possibly related to increases in cellular proliferation and differentiation, there was also evidence for increases in large molecule synthesis. For example, RNA metabolism (“positive regulation of RNA biosynthetic process”; “positive regulation of transcription by RNA polymerase II”) and ‘macromolecule synthesis” were affected by treatment. Less significant but still noteworthy were pathways related to behavior and stress responses. The only pathway showing a significant enrichment was the “AP-1 transcription factor network” that includes genes responsive to external stimuli (Fos, Fosb, Dusp1, Junb, Th, Ccn1 and Agt). GO:Molecular Function terms were mostly related to transcription factor activity and RNA transcription, which is consistent with the correlations between ENHO expression and ribosomal genes in open access transcriptome data (Fig. 7A).
Adropin improves spatial learning and memory in 18-month-old B6 mice
Tests of spatial learning and memory and novel object recognition (NOR) were used to determine whether adropin has a positive effect on cognitive function in 18-month-old male B6 mice. Male AdrTG mice performed significantly better in tests of spatial learning and memory, and of recognition memory compared to age-matched WT controls (Fig. 8D–E). Treatment with adropin34-76 similarly improved performance in trials (Fig. 8D–E). Fasting glucose and insulin data were recorded at the completion of the study; no significant treatment effects were observed for either parameter suggesting that the behavioral effects are independent of marked improvements in glycemic control (Fig. S5B). Collectively, these results indicate that high expression of adropin may protect against mild cognitive decline with aging, and that acute treatment may reverse the effects of aging.
AdrTG mice exhibit improved resilience in tests of energy balance and ambulatory activity
We next compared how adropin over expression affects aging-related changes of whole-body energy balance and ambulatory movement using mice either ‘middle aged’ (M, 8–10 mo) or ‘old’ (O, 16–20 mo.). The respiratory exchange ratio (RER, VCO2/VO2) provides an estimate of whole body substrate selection68. The RER increases during the dark period as chow-fed mice move to oxidizing ingested carbohydrates and relying less on mobilized fatty acid (Fig. S6A, B). WT-O mice exhibited low RER, indicating increased dependency on fat reserves (Fig. S6A). In females, the RER transition between light and dark periods was more pronounced in AdrTG, irrespective of age (Fig. S6B). Oxygen consumption exhibited the diurnal rhythm predicted from increased ambulatory activity and food intake during the dark period (Fig. S6C, D). This feature of energy metabolism was not markedly affected by genotype.
Food intake and changes in body weight during the recordings were consistent with RER data, indicating improved maintenance of body weight in M and old-AdrTG mice. Male WT-O mice lost weight during the recording (mean ± SD and n for weight gain/loss in g, young-WT −0.1 ± 0.2, n = 4; old-WT −5.8 ± 3.4, n = 3; young-AdrTG −0.4 ± 1.0, n = 4; old-AdrTG −1.7 ± 0.5, n = 3). Female WT mice also tended to lose more weight (mean ± SD for weight gain/loss in g, young-WT −3.0 ± 2.5; old-WT −1.6 ± 2.9; young-AdrTG −0.1 ± 0.5; old-AdrTG −0.6 ± 0.5, n = 4 all groups). Weight loss correlated with lower food intake during the recording period (Fig. S6E). Pooling data from male and female mice indicated WT mice were more prone to weight loss during recordings (−2.4 ± 3.0 vs. −0.6 ± 0.8, p < 0.05). Advanced age may exacerbate the phenotype (Fig. S6F).
Ambulatory movement exhibited the predicted diurnal profile, irrespective of age or sex or genotype (Fig. S6G, H). Aging of WT mice, but not in AdrTG mice, appeared to associate with reduced movement (Fig. S6G-I). A negative energy balance indicated by low food intake and weight loss might be predicted to account for this phenotype. Food intake is a strong predictor of weight gain/loss (Fig. S7A). However, it is not a strong predictor of ambulatory movement (Fig. S7B). Weight gain/loss is also only a weak predictor of ambulatory movement (Fig. S7C). AdrTG aged < 6 months exhibit a lean phenotype and improved glycemic control14. At 18 months of age, evidence for modest differences in nutrient partitioning that suggest a lean phenotype are still observed (Fig. S8A-F), however glucose clearance is normal (Fig. S8G).
Collectively, these results indicate significant genotype effects on feeding behavior, substrate selection glucose vs. fat oxidation and ambulatory movement. As prolonged anorexia is not compatible with survival, superior adaptation of AdrTG to the stress of being moved to a novel environment (metabolic caging) is a plausible explanation for the behavioral and metabolic phenotypes.
Discussion
These results indicate that adropin functions support brain health during aging. In the human and mouse brain transcriptome, expression of the ENHO transcript encoding adropin correlates positively with gene networks involved in mitochondrial energy metabolism and the synthesis of macromolecules supporting cellular functions. The brain accounts for 20% of the body’s energy consumption, with glucose being the primary fuel source69. It is therefore tempting to speculate that adropin expression in the brain will positively correlate with glucose uptake and utilization. Certainly, the positive correlation with processes involved in macromolecule synthesis and RNA metabolism suggests expression associates positively with metabolic activity at a cellular level.
The inverse associations with AKT3 and GSK3B in the human brain indicate this interpretation is perhaps overly simplistic. AKT3 belongs to a family of serine/threonine kinases (AKT1–3), and regulates brain development and cognitive function70. Phosphorylation of GSK3 by AKT3 may also affect behavior70. Given the roles of GSK3 in mediating tau hyperphosphorylation47 and affecting behavior, the inverse association with GSK3B is also potentially of interest. Further experiments investigating the regulation of glucose metabolism in astrocytes and neurons, and of the relationships between adropin and GSK3 activity in the brain, are clearly needed.
Peak adropin expression in the first decade of life suggests adropin has a role in regulating developmental processes. This hypothesis is supported by the phenotype of adropin knockout mice which display deficits in synapse formation, coupled with decreased locomotor activity and impaired motor coordination31. We also show that adropin at physiological concentrations acts directly on neurons, significantly enhancing the number, length, and thickness of neurites in the early development of cultured neurons (within first 2 days in culture). Increased thickness of primary neurites correlates with increased neuronal surface area for housing ionic channels/receptors, promoting efficiency of neuronal conductivity and synaptic transmission. In addition, adropin significantly increases neuronal activity in primary hippocampal neurons. Together, the neuritogenic and neuroexcitatory effects of adropin on hippocampal neurons may contribute to its function in promoting high cognition in animals and humans.
Growth and development of the human brain requires an abundance of calorie-dense diets, including essential fatty acids to provide energy and substrates for cell division, cellular morphogenesis, and synaptic functions71. It is therefore interesting to note the positive correlations between circulating adropin levels and selection of diets with high energy content72,73. Nutrient-sensing mechanisms have been identified that drive macronutrient preferences to match macronutrients selection with energy requirements74,75. While speculative, preference for high fat diets in people with high adropin levels could correlate with increased nutrient demand by the nervous system. Indeed, in young people plasma adropin concentrations appear to correlate positively with a lean phenotype41. Further investigation of the relationships between adropin expression, plasma adropin concentrations, and nutrient requirements of the brain are clearly needed.
The relationships between ENHO and the expression of candidate genes (APOE, CLU/ApoJ) and pathways involved in cholesterol metabolism in the brain are relevant to aging. APOE has a critical role in mediating cholesterol transport in the brain43. CLU/ApoJ has also been implicated in lipid metabolism, and functions as extracellular chaperone linked to Aβ clearance and toxicity45. The negative relationship between ENHO expression and “REACTOME_CHOLESTEROL_BIOSYNTHESIS” is also consistent with our recent studies showing relationships between adropin and cholesterol metabolism. In humans and NHPs we have observed an inverse correlation between adropin and plasma markers of cholesterol metabolism28,41. It is reasonable to speculate that high adropin expression in the brain could increase plasma concentrations of adropin peptide, and correlate with brain cholesterol metabolism. Plasma adropin concentrations in humans are higher in males compared to females, particularly early in life, and decline with aging41. Based on gene expression analysis, neural tissues appear to be a plausible site of origin for circulating adropin in humans. It will be important to determine whether adropin protein levels correlate with gene expression. Expression of the mature Enho mRNA correlates with levels detected in ribosomal fractions in the mouse liver. Whether this relationship is also observed in the nervous system needs further study.
The conservation of the correlations between adropin and metabolic pathways between mice and humans suggests that the responses of mice to treatment could be translatable. It is important to note that the brain samples in the clinical studies came from individuals who died of natural causes or by accident. In contrast, the mice were euthanized at specific ages and times. This latter point is potentially important, as ENHO expression in the NHP brain exhibits a circadian profile28.
In humans, advanced age appears to result in changes in the pathways correlating with ENHO expression. Correlations observed in those with advanced age are also dependent on dementia status. In the ‘old-old’ without dementia, correlations develop with gene networks involved in morphogenesis, including the growth of axonal and synaptic processes. Adropin has been implicated in brain development in mice31, and here we observed potent effects of synthetic adropin on development of neuronal processes in cultured neurons, indicating a direct effect. This could suggest adropin signaling maintains neuronal processes critically involved in cognitive function during aging.
In ‘old-old’ people diagnosed with dementia, the ENHO transcript clustered with gene networks involved in mitochondrial energy metabolism, with terms related to vascular function also appearing in the analysis. Adropin has been shown to regulate differentiation of human umbilical vein endothelial cells into capillary-like structures, while transgenic over expression of adropin stimulated angiogenesis and improved blood flow in a mouse model of ischemia20. Clinical studies have observed positive correlations between circulating adropin levels and endothelial function57,76. Actions of adropin on vascular function could be relevant to the aging brain, as reduced cerebrovascular blood flow has been linked to aging-related cognitive impairment and dementia77.
The results from studies using aged mice provide further indication of high adropin expression benefiting cognitive performance. Furthermore, providing synthetic adropin as a supplement to aged mice in which adropin protein levels have declined appears to have a therapeutic effect. However, the positive correlations between ENHO expression and protein markers of tau pathology contradict this conclusion. There is considerable variation in signatures of brain inflammation; these markers do not always correlate with dementia status in the ‘old-old’ cohort used for the current study48.
It is difficult to interpret the differences between the correlative data observed in postmortem samples from people aged from birth to 40 years (mature adults) and the ‘old-old’. Reduced fidelity in transcriptional and posttranscriptional mechanisms affecting transcript abundance is a plausible explanation for lower correlation coefficients. However, survivorship bias should be considered when comparing differences in pathways and biological processes correlating with the ENHO transcript in the D and ND groups. Transitioning to correlations with neural morphogenesis could indicate a survival advantage in the ‘old-old’ without dementia. A similar reasoning could be applied to the retention of mitochondrial processes and transitioning to associations with vascular function in the ‘old-old’ with dementia. This does not necessarily imply that high levels of adropin expression per se confers a survival benefit. Rather, retaining regulatory control of gene networks that include the ENHO gene may act in concert to confer a survival advantage in the old-old. On the other hand, the data from mouse studies clearly imply that high adropin expression may be beneficial.
Similarly, the trend for higher ENHO expression in the ‘old-old’ carriers of the APOE ε4 allele could indicate survivor bias. GWAS have linked APOE gene variants to longevity78. Inheritance of the ε4 allele increases risk of early mortality from cardiovascular disease and LOAD. That ENHO expression tends to be higher in people aged over 75 y with >0 APOE e4 alleles could indicate a selection advantage. In this scenario, a combination of the APOE e4 allele with high adropin expression delays the development of LOAD. An alternative interpretation is that inheritance of APOE ε4 allele associates with cellular stress phenotypes in the nervous system that increase ENHO expression. For example, increases in Enho expression in the mouse live have been linked to oxidative stress30. The two theories are also not mutually exclusive, with activation of adropin expression and signaling in response to stress having a protective effect.
Aging-related transitions in gene networks that correlate with ENHO could also indicate responses to changes in the cellular environment. Cellular stress responses driving age-related inflammation could dominate, thereby altering the regulatory framework driving the transcriptional and posttranscriptional processes affecting the levels of ENHO and other transcripts. The observation that the gene networks correlating with ENHO differ between the ‘old-old’ diagnosed with dementia (mostly LOAD) and those who died with normal cognitive function is consistent with this theory.
It is important to note some of the limitations of the human studies. For example, differences in nutrition and environment during childhood between participants of the GTEx and the Aging, Dementia and TBI Study could be important. The participants in the latter study were born in the early 20th Century when obesity and type 2 diabetes were less prevalent. On the other hand, postmortem samples in the GTEx portal come from people living in an obesogenic environment.
Pending clinical studies designed to administer adropin to people with advanced age, the human data reported here are by necessity correlative. Mouse models were used to investigate whether higher adropin expression confers an advantage in aging, and also whether administration of synthetic peptide would be beneficial.
The AdrTG mice used for these studies exhibit increased adropin protein expression in the brain. However, the use of a ubiquitous promoter increases expression in all tissues14,17,30,41. The results from experiments using 18-month-old male mice nevertheless suggest increasing expression of adropin improves cognitive function. Cognitive function was assessed by tests of spatial learning and memory and NOR. The use of mice over expressing adropin throughout the lifespan complicates the interpretation of these results. However, injections of adropin peptide improved cognitive performance in 18-month-old B6 mice. The effects of adropin to improve cognitive performance can thus be dissociated from metabolic phenotypes earlier in life. Moreover, the results from this acute treatment experiment suggest that treatment with synthetic adropin can reverse aging-related cognitive impairment.
The results from assessing whole body energy balance could be interpreted as indicating increased resilience in response to a novel environment. Old mice transferred to metabolic cages exhibit anorexia, reduced activity, weight loss and mobilization of fatty acids. This response was not evident in age-matched AdrTG. Future studies examining how adropin over expression affect behavioral flexibility and stress responses in aging are clearly warranted. While technically challenging, behavioral studies using ‘old-old’ mice aged >2 years could also be informative.
A weakness with the mouse studies is the lack of apparent clear mechanism explaining the phenotype. Gene expression analysis suggests reduced inflammation and neurotrophic mechanism as potential factors. Other mechanisms based on the known functions of adropin are possible that include effects on blood flow20 or the blood brain barrier59. Direct neurotrophic actions on neurons were also observed. However, it is currently not known whether adropin34-76 administered ip. penetrates the blood brain barrier.
The current results nevertheless provide an indication that high expression during the later stages of life, or administration of the synthetic peptide or derivative thereof, confer a benefit. Further experiments exploring mechanism(s) are clearly indicated. Whether adropin expression correlates with lifespan, and whether over expression is effective in mouse models of severe LOAD, also needs to be explored.
In summary, current demographic trends indicate that the burden of dementia will increase this century as the population ages. The current study has applied a translational approach to identify a novel candidate for developing therapies to improve cognitive function in aging. Further investigation of the adropin neuropeptide with respect to cognitive function and brain health are clearly warranted.
Methods
Analysis of transcriptome data
For pre-processing of The Aging, Dementia and TBI Study Patient, datasets were downloaded from https://aging.brain-map.org/download/index. The data included: (i) de-identified clinical information for 107 patient donors, (ii) normalized gene-level FPKM (Fragments Per Kilobase of transcript per Million mapped reads) expression for 377 unique samples obtained from these donors, (iii) A sample metadata table and (iv) a gene ID and gene symbol table for 50,281 unique genes. To determine whether the samples were normalized, we plotted the density of histograms for all samples. To perform the correlation analysis, we pre-processed the expression matrix using “log2(FPKM+1)”. All analyses were performed using Bioconductor79,80 in R.
GSEA (Genome-wide gene set enrichment analysis) used a scaled correlation matrix to reveal ENHO-coregulated gene pathways81. GSEA is used to evaluate the enrichment or depletion of a given gene set/pathway relative to one of the two functional states, which are distantly connected by a gradient of genes pre-ranked based on a measurable activity. Typically, these are differentially expressed genes. In our analysis, we defined the two states as, (i) a gene state positively associated with ENHO expression, and (ii) a gene state antagonistic or incompatible with ENHO expression.
To perform GSEA, 5529 experimentally-verified biological pathways were downloaded from the Molecular Signature Database82 (V7.1 C2) (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). Pearson Correlation coefficients were calculated between ENHO and each of the 50281 genes. This was done using the entire dataset containing all 377 samples or only a portion of them with specific features (Dementia/No Dementia, Male/Female or both). The correlation coefficients were then ranked, centered and scaled. The resulting pre-ranked correlation coefficients were used to search for enriched or depleted pathways from the 5529 experimentally-verified biological pathways using the “fgsea” package in R. Genes not included in the MSigDB pathways were removed from the list of pre-ranked genes. Significant pathways with an adjusted p value ≤ 0.05 were selected for further analyses. Top 20 enriched or depleted pathways were plotted. These significant pathways were saved into excel files.
For the other datasets referenced in the Result sections, data were downloaded and log transformed if needed. Genes were ranked by the correlation coefficient determined using Microsoft Excel. Functional enrichment analysis used the ToppFun tool (https://toppgene.cchmc.org). Biological processes in separate populations (D, ND, age < 40 y) were assessed using the PercaiAI CompBio platform; the Assertion Engine was then used to determine conservation of pathways between populations. Each Assertion produces similarity scores from 0 to 1, with scores >0.1 indicating substantial overlap. Outputs of Assertion Engine queries are heatmaps that show overlap scores between datasets.
Neuronal cell culture, phase contrast imaging, and ImageJ neurite tracing analysis
Hippocampal neurons were dissected from B6 mice and cultured from newborn pups (postnatal day 2, P2) as described previously83,84. Briefly, hippocampal tissue was isolated and single cells were dissociated with papain (50 µg/mL). Single-cell suspensions were created via trituration with pipettes of decreasing size. The cells were diluted in culture media (neurobasal medium, 2% B27, L-Glutamine [200 mM], 4% FBS, and penicillin-streptomycin [Invitrogen]) and plated on dishes coated with poly-D-lysine (100 µg/mL) and laminin (2 µg/mL). For phase-contrast imaging, cells were cultured on glass-bottom petri dishes. For neuronal activity recording, primary cultured neurons were cultured on Multi-electrode array (MEA) Neurochip (Multi Channel Systems) was coated with poly-D-lysine and laminin, and cells were plated directly on the Neurochip. Hippocampal neurons were kept at 37 °C in an airtight modular incubator chamber (Thermo-Forma) circulated with 5% carbon dioxide and medical air. Every 3–4 days, half of the culture media was replaced with fresh media.
To examine the effect of adropin on the formation of early networks, culture media supplemented with 1 nM, 10 nM, or 100 nM adropin was added to culture dishes on the same day as plating (day 0). We counted and measured the number of branches and total neurite outgrowth (measured in each phase contrast image taken) on day 2 and the neuritic thickness of primary neurites (processes derived directly from cell bodies) on day 14. The number and length of neurites in day 2 and the primary neurites in day 14 cultures are easily discernible, and hence we chose to analyze these parameters. Quantitative data and statistical analyses are shown in Fig. 5. Hippocampal neurons grew for 2 days (day 2), and phase contrast images were taken with an inverted microscope (Olympus CKX53). To examine adropin effects on developed neuronal networks, primary hippocampal neurons were grown for 9 days in normal neurobasal culture media. On day 9, 1 nM, 10 nM, or 100 nM adropin was added to culture media and phase contrast images were taken 5 days later (day 14). In all phase contrast experiments, control dishes received no adropin; microscope parameters were the same for control and treatment dishes.
ImageJ was used to analyze phase contrast images of primary hippocampal cells. The ImageJ plugin NeuronJ was employed to measure neurite characteristics in phase contrast images of hippocampal neurons 2 or 14 days in culture in control (no adropin added) and adropin- (1, 10, and 100 nM) added culture as previously described61,62,84,85. NeuronJ was programmed to output total neurite outgrowth (µm) and the number of neuritic branches for day 2 cultures and the thickness of primary neurites in day 14 cultures. This is because in day 2 cultures, the length and number of neurites from each cell can be easily identified and traced using ImageJ. For cells that were maintained in culture for 14 days, it is hard to judge the terminal end of each neurite, as extensive networks are formed and glial cells are proliferated and formed layers underneath. We, therefore, focused on analyzing the thickest primary neurites in day 14 cultures. In addition, for day 2 neuritic outgrowth and branching analyses, all neurites within a phase contrast image were analyzed. For day 14 neuritic thickness analysis, the five thickest primary neurites were measured in each image.
Multi-electrode array (MEA) recording of neuronal activity and spiking frequency analysis
The MEA2100-System Neurochip consists of 60 electrodes organized in an 8*8 grid, designed to capture neuronal electrical activity (Multi Channel Systems). Hippocampal neurons were cultured on the Neurochip as described above and grown for nine days (day 9) to allow for synapse formation. Neuronal activity was measured with the MEA2100-Lite head stage connected to the MCS-IFB interface board. The baseline of neuronal activity was first recorded for 4 min or until the neuronal activity among majority of electrodes was stabilized to act as a pre-treatment baseline. After 4 min, 100 nM adropin was added to the medium, and electrical activity recordings were taken for 8 more minutes by the Multichannel Experimenter software. Neuronal activity was quantified as “spikes per minute,” the number of neuronal firings that occurred within 60 s of recording. Spikes per minute (pre- and post-adropin addition) was normalized to baseline spikes per minute (pre-adropin) for each electrode. This allowed a fair comparison between all electrodes, whether they were previously quiescent or actively firing during baseline recordings.
Animal studies
Studies involving mice were conducted with the approval of the Institutional Animal Care and Use Committees at the Saint Louis University School of Medicine and the University of Florida. Sentinels were tested regularly to ensure our facility is virus- and pathogen- free. Food and water were available on an ad libitum basis and the rooms had a 12 h light-dark cycle with lights on at 0600 h.
AdrTG were generated and maintained as previously described14. For aging studies, mice were maintained in group housing (3–4/cage) with ad libitum access to water and standard rodent chow. For the experiment investigating the response of aging mice to adropin peptide, male B6 mice were purchased at 18 months of age from Jackson Laboratories (Bar Harbor, ME). Mice were acclimated to local conditions for 2 week and then divided into two weight matched groups (n = 15/group). The mice were treated with 90 nmol/kg/d adropin34-76 (ABClonal Science, 98.66% purity) suspended in 0.9% sterile saline with 0.1% BSA. The peptide was resuspended daily prior to injection and administered as a single ip. injection (0.1 ml volume) at 0900 h. Controls received saline diluent only. Mice were weighed daily; after 2 weeks of injections, mice were subjected to behavioral testing. Treatment continued during behavioral testing.
At completion, mice were fasted for 6 h. Blood glucose (tail snip) levels were recorded using a glucometer; blood samples were collected for analysis of measurement of insulin by ELISA (Ultra Sensitive Mouse Insulin ELISA kit, Crystal Chem USA, IL, USA). Brain tissue samples were collected and snap frozen using liquid nitrogen. Fasting glucose and glucose clearance were assessed in mice fasted for 6 h. Glucose tolerance tests were performed using 1 mg/kg dextrose administered intraperitoneally (ip.), as previously described30.
To visualize cells expressing adropin, an IRES-Cre was inserted into the 3′untranslated region (UTR) of the adropin open reading frame in exon 2. The targeting vector was constructed using recombineering system. Isogenic DNA containing the Enho locus was retrieved from genomic colony RP23–100C7 of C57Bl/6 BAC genomic library via gap repair. An IRES-Cre-Frt-neo-Frt was inserted into 3′ 53 bp downstream of the translational stop codon in exon 2. For gene targeting, 50 μg of linearized targeting vector consisting of 3.5 kb 5′arm and 7.2 kb 3′ arm was electroporated into Bruce4 B6 embryonic stem (ES) cells. Homologous recombination in targeted clones was confirmed with Fidelity PCR at the 5′ and 3′ ends. The fragments produced from Fidelity PCR with these primers were sequenced to verify the correctness of recombination.
Correctly targeted ES cells were injected into Albino B6 blastocysts; germline transmitting chimeric mice were obtained and mated with Albino B6 mice to generate heterozygous carriers of the EnhoIRES-Cre-Frt-neo-Frt on the B6 background. The Frt-neo-Frt sequence was removed using B6;SJL-Tg(ACTFLPe)9205Dym/J transgenic mice purchased from the Jackson laboratory. EnhoIRES-Cre mice were then crossed onto the B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J strain in which a loxP-flanked stop cassette prevents transcription of a red fluorescent protein variant (tdTomato) driven by a CAG promoter (5).
Immunohistochemistry
Surgical anesthesia was achieved using intraperitoneal administration of a cocktail of Ketamine (100 mg/kg) and Xylazine (10 mg/kg), 12 weeks-old EnhoIRES−Cre;ROSA-tdTomato mice were sacrificed by intra-aortic perfusion of 5 ml of ice-cold PBS followed by 50 ml of ice-cold paraformaldehyde 4%. The brain was then removed from the skull, post-fixed overnight in the same fixative, cryoprotected in 30% sucrose solution before being snap freeze in isopentane (−40 °C). Frozen brains were then cut using a cryostat (CM1950, Leica) to collect 30 μm serial sections in PBS.
Immunohistochemistry (IHC) was performed on free-floating sections. To assess Adropin expression in post-mitotic neurons, astrocytes, and endothelial cells antibodies against either NeuN (Abcam cat. # EPR12763; ;1:500 dilution), GFAP (Sigma HPA056030; dilution 1:1000), CD31 (Novus Biologicals NB-80639; 1:100) were used respectively. Briefly, after 1 h in PBS containing 10% Normal Goat Serum (NGS) and 0.2% Triton X100, sections were incubated for 24 h at 4 °C in PBS containing 1% NGS, 0.2% Triton X100 and primary antibody. Goat-Alexa 488 conjugated anti-rabbit IgG (Invitrogen A32731;1:400) was used as secondary antibody. Sections were then mounted on slides and cover slipped using FluorSave reagent (Calbiochem). tdTomato and Alexa 488 fluorescence were visualized with an Olymups confocal using the Fluorview1000 software.
RNA sequencing and analysis
Total RNA integrity was determined using Agilent Bioanalyzer or 4200 Tapestation. Library preparation was performed with 5–10 ug of total RNA with a Bioanalyzer RIN score >8.0. Ribosomal RNA was removed by poly-A selection using Oligo-dT beads (mRNA Direct kit, Life Technologies). mRNA was then fragmented in reverse transcriptase buffer and heating to 94 degrees for 8 min. mRNA was reverse transcribed to yield cDNA using SuperScript III RT enzyme (Life Technologies, per manufacturer’s instructions) and random hexamers. A second strand reaction was performed to yield ds-cDNA. cDNA was blunt ended, had an A base added to the 3′ ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments were then amplified for 12–15 cycles using primers incorporating unique dual index tags. Fragments were sequenced on an Illumina NovaSeq-6000 using paired end reads extending 150 bases.
Samples were prepared according to library kit manufacturer’s protocol, indexed, pooled, and sequenced on an Illumina NovoSeq. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq software and a custom python demultiplexing program with a maximum of one mismatch in the indexing read. RNA-seq reads were then aligned to the Ensembl release 76 primary assembly with STAR version 2.5.1a1. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.6-p52. Isoform expression of known Ensembl transcripts were estimated with Salmon version 0.8.23. Sequencing performance was assessed for the total number of aligned reads, total number of uniquely aligned reads, and features detected. The ribosomal fraction, known junction saturation, and read distribution over known gene models were quantified with RSeQC version 2.6.24.
All gene counts were then imported into the R/Bioconductor package EdgeR5 and TMM normalization size factors were calculated to adjust for samples for differences in library size. Ribosomal genes and genes not expressed in the smallest group size minus one sample greater than one count-per-million were excluded from further analysis. The TMM size factors and the matrix of counts were then imported into the R/Bioconductor package Limma6.
T-Maze training and testing procedures, novel object recognition
Behavioral experiments were conducted between 0730 and 1400 h. The T-maze is both a learning task based on working-memory and a reference-memory task. The T-maze consisted of a black plastic alley with a start box at one end and two goal boxes at the other. The start box was separated from the alley by a plastic guillotine door that prevented movement down the alley until raised at the onset of training. An electrifiable floor of stainless-steel rods run throughout the maze to deliver a mild scrambled foot-shock.
Mice were not permitted to explore the maze prior to training. A block of training trials began when a mouse was placed into the start box. The guillotine door was raised and a cue buzzer sounded simultaneously; 5 s later, foot-shock was applied. The arm of the maze entered on the first trial was designated “incorrect” and the mild foot-shock was continued until the mouse entered the other goal box, which in all subsequent trials was designated as “correct” for the particular mouse. At the end of each trial, the mouse was returned to its home cage until the next trial.
Mice were trained until they made one avoidance. Training used an inter-trial interval of 60 s, the buzzer was of the door-bell type sounded at 55 dB, and shock was set at 0·35 mA (Coulbourn Instruments scrambled grid floor shocker model E13–08). Retention was tested 1 week later by continuing training until mice reached the criterion of five avoidances in six consecutive trials. The results were reported as the number of trials to criterion for the retention test.
Novel object recognition (NOR) was tested the 5 days following T-maze retention testing. NOR is a declarative memory task that involves the HIP when, as performed here, the retention interval is 24 h after initial exposure to the objects86. Mice were habituated to an empty apparatus for 5 min a day for 3 days prior to entry of the objects. During the training session, the mouse was exposed to two identical objects which it was allowed to examine for 5 min. The apparatus and the objects were cleaned between each mouse. 24 h later, the mouse was exposed to one of the original objects and a new novel object in a new location and the amount of time spent examining each object was recorded. The novel object was made from the same material as the original object and of the same size, but a different shape. This eliminated the possibility of smell associated with a particular object being a factor. The underlying concept of the task is based on the tendency of mice to spend more time exploring new, novel objects than familiar objects. The greater the retention/memory at 24 h, the greater the discrimination index (DI). The time with new object (tn) and time spent with the old object (to) was used to calculate the DI [DI = (tn-to)/(tn+to)]87.
Measurements of gene and protein expression
Expression of candidate genes was assessed using qRT-PCR30. The extraction of the total RNA from was performed by using kits from Applied Biosystems. cDNA was synthesized with cDNA reverse transcription kit from Quanta. PCR was conducted using a QuantStudio Realtime PCR machine, Applied Systems using primers from Integrated DNA Technology. Data were normalized using three reference genes (HPRT1, 36B4, PPIB). A list of primers and their sequence is provided in Table S1. Quantitative PCR was performed in 384-well plates using SYBR Green and QuantStudio 7 Detection Systems (Applied Biosystems, Life Technologies).
Adropin peptide levels in brain tissues lysate were assessed by Western blot using a mouse monoclonal antibody (Cayman Chemical cat. no. 14117, Ann Arbor, MI; 1:1000 dilution)40. An HSP90 rabbit polyclonal antibody (Cell Signaling Technology cat. no. 4874, Danvers, MA; 1:1000 dilution) was used as a reference for loading.
Statistics
Mouse data were analyzed using Microsoft Excel or Graph Pad Prism software; differences between control and treatment groups were assessed by Student’s t test. One-way analysis of variance (ANOVA) and Tukey’s HSD post hoc tests were used to analyze significant differences between control and the three concentrations of adropin for neuritic outgrowth, number of branches, and thickness of primary neurites. Values were considered statistically significant at the level of p < 0.05. The data are presented as mean ± S.E.M. Each experiment was replicated a minimum of three times; the actual N and p values were provided in the text or figure legend.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
RNA-sequencing data from the experiment investigating the response of 18-month-old C57BL/6J mice to adropin peptide is available from the Gene Expression Omnibus (GSE179088). RNA-sequencing data used to compare ENHO expression in the brain of wild type and transgenic mouse models of dementia were from the Gene Expression Omnibus (GSE125957). Human expression data are available from the Atlas of the Developing Human Brain (www.brainspan.org), Genotype-Tissue Expression (GTEx) portal (https://gtexportal.org/home/), and The Aging, Dementia and TBI Study (http://aging.brain-map.org/). Results from our analysis of the “old-old” dataset downloaded from http://aging.brain-map.org/ are available online (http://pharmacology.slu.edu/results/andrew/TBI_Dementia/). An interactive shiny serve is also available at http://pharmacology.slu.edu/shiny/tbi_shiny/.
Code availability
No code was generated for this paper.
References
Korkki, S. M., Richter, F. R., Jeyarathnarajah, P. & Simons, J. S. Healthy ageing reduces the precision of episodic memory retrieval. Psychol. Aging 35, 124–142 (2020).
Langnes, E. et al. Anterior and posterior hippocampus macro- and microstructure across the lifespan in relation to memory-A longitudinal study. Hippocampus 30, 678–692 (2020).
Tromp, D., Dufour, A., Lithfous, S., Pebayle, T. & Despres, O. Episodic memory in normal aging and Alzheimer disease: insights from imaging and behavioral studies. Ageing Res Rev. 24, 232–262 (2015).
Petersen, R. C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 256, 183–194 (2004).
Bennett, D. A. et al. Natural history of mild cognitive impairment in older persons. Neurology 59, 198–205 (2002).
Morley, J. E. Mild cognitive impairment-a treatable condition. J. Am. Med. Dir. Assoc. 15, 1–5 (2014).
Stefaniak, J. & O’Brien, J. Imaging of neuroinflammation in dementia: a review. J. Neurol. Neurosurg. Psychiatry 87, 21–28 (2016).
Chandra, A. et al. Applications of amyloid, tau, and neuroinflammation PET imaging to Alzheimer’s disease and mild cognitive impairment. Hum. Brain Mapp. 40, 5424–5442 (2019).
Bradburn, S., Murgatroyd, C. & Ray, N. Neuroinflammation in mild cognitive impairment and Alzheimer’s disease: a meta-analysis. Ageing Res. Rev. 50, 1–8 (2019).
Crimmins, E. Future Directions for the Demography of Aging: Proceedings of a Workshop (eds M. K. Majmundar & M. D. Hayward) pp 3–30 (The National Academies Press, Washington, DC, 2018).
Makarewich, C. A. & Olson, E. N. Mining for Micropeptides. Trends Cell Biol. 27, 685–696 (2017).
Andrews, S. J. & Rothnagel, J. A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15, 193–204 (2014).
Clark, H. F. et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genom. Res. 13, 2265–2270 (2003).
Kumar, K. G. et al. Identification of adropin as a secreted factor linking dietary macronutrient intake with energy homeostasis and lipid metabolism. Cell Metab. 8, 468–481 (2008).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Wang, P. et al. Mapping spatial transcriptome with light-activated proximity-dependent RNA labeling. Nat. Chem. Biol. 15, 1110–1119 (2019).
Gao, S. et al. Regulation of substrate oxidation preferences in muscle by the peptide hormone adropin. Diabetes 63, 3242–3252 (2014).
Gao, S. et al. Therapeutic effects of adropin on glucose tolerance and substrate utilization in diet-induced obese mice with insulin resistance. Mol. Metab. 4, 310–324 (2015).
Gao, S. et al. The peptide hormone adropin regulates signal transduction pathways controlling hepatic glucose metabolism in a mouse model of diet-induced obesity. J. Biol. Chem. 294, 13366–13377 (2019).
Lovren, F. et al. Adropin is a novel regulator of endothelial function. Circulation 122, S185–S192 (2010).
Thapa, D. et al. Adropin regulates pyruvate dehydrogenase in cardiac cells via a novel GPCR-MAPK-PDK4 signaling pathway. Redox Biol. 18, 25–32 (2018).
Thapa, D. et al. Adropin reduces blood glucose levels in mice by limiting hepatic glucose production. Physiol. Rep. 7, e14043 (2019).
Thapa, D. et al. Adropin treatment restores cardiac glucose oxidation in pre-diabetic obese mice. J. Mol. Cell Cardiol. 129, 174–178 (2019).
Stein, L. M., Yosten, G. L. & Samson, W. K. Adropin acts in brain to inhibit water drinking: potential interaction with the orphan G protein-coupled receptor, GPR19. Am. J. Physiol. Regul. Integr. Comp. Physiol. 310, R476–R480 (2016).
Rao, A. & Herr, D. R. G protein-coupled receptor GPR19 regulates E-cadherin expression and invasion of breast cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1318–1327 (2017).
Altamimi, T. R. et al. Adropin regulates cardiac energy metabolism and improves cardiac function and efficiency. Metabolism 98, 37–48 (2019).
Butler, A. A. et al. Low circulating adropin concentrations with obesity and aging correlate with risk factors for metabolic disease and increase after gastric bypass surgery in humans. J. Clin. Endocrinol. Metab. 97, 3783–3791 (2012).
Butler, A. A. et al. Low plasma adropin concentrations increase risks of weight gain and metabolic dysregulation in response to a high-sugar diet in male nonhuman primates. J. Biol. Chem. 294, 9706–9719 (2019).
Ganesh Kumar, K. et al. Adropin deficiency is associated with increased adiposity and insulin resistance. Obes. (Silver Spring) 20, 1394–1402 (2012).
Banerjee, S. et al. Hepatocyte expression of the micropeptide adropin regulates the liver fasting response and is enhanced by caloric restriction. J. Biol. Chem. 295, 13753–13768 (2020).
Wong, C. M. et al. Adropin is a brain membrane-bound protein regulating physical activity via the NB-3/Notch signaling pathway in mice. J. Biol. Chem. 289, 25976–25986 (2014).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 13, 397–406 (2014).
Mure, L. S. et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359, eaao0318 (2018).
Consortium, G. T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013).
O’Dowd, B. F. et al. A novel gene codes for a putative G protein-coupled receptor with an abundant expression in brain. FEBS Lett. 394, 325–329 (1996).
Hoffmeister-Ullerich, S. A., Susens, U. & Schaller, H. C. The orphan G-protein-coupled receptor GPR19 is expressed predominantly in neuronal cells during mouse embryogenesis. Cell Tissue Res. 318, 459–463 (2004).
Alexander, S. P. et al. Class A Orphans (version 2019.5) in the IUPHAR/BPS Guide to Pharmacology Database. IUPHAR/BPS Guide Pharmacol CITE 5, 52 (2019).
Foster, S. R. et al. Discovery of Human Signaling Systems: pairing Peptides to G Protein-Coupled Receptors. Cell 179, 895–908 e821 (2019).
Chatterjee, M., Schild, D. & Teunissen, C. E. Contactins in the central nervous system: role in health and disease. Neural Regen. Res 14, 206–216 (2019).
Yang, C., DeMars, K. M. & Candelario-Jalil, E. Age-Dependent Decrease in Adropin is Associated with Reduced Levels of Endothelial Nitric Oxide Synthase and Increased Oxidative Stress in the Rat Brain. Aging Dis. 9, 322–330 (2018).
Ghoshal, S. et al. Adropin: an endocrine link between the biological clock and cholesterol homeostasis. Mol. Metab. 8, 51–64 (2018).
Zhang, Y. et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53 (2016).
Lanfranco, M. F., Ng, C. A., & Rebeck, G. W. ApoE Lipidation as a Therapeutic Target in Alzheimer’s Disease. Int. J. Mol. Sci. 21, 6336 (2020).
Yamazaki, Y., Zhao, N., Caulfield, T. R., Liu, C. C. & Bu, G. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 15, 501–518 (2019).
Foster, E. M., Dangla-Valls, A., Lovestone, S., Ribe, E. M. & Buckley, N. J. Clusterin in Alzheimer’s Disease: mechanisms, Genetics, and Lessons From Other Pathologies. Front. Neurosci. 13, 164 (2019).
Miao, L. et al. mTORC1 is necessary but mTORC2 and GSK3beta are inhibitory for AKT3-induced axon regeneration in the central nervous system. Elife 5, e14908 (2016).
Hernandez, F., Lucas, J. J. & Avila, J. GSK3 and tau: two convergence points in Alzheimer’s disease. J. Alzheimers Dis. 33, S141–S144 (2013). Suppl 1.
Miller, J. A. et al. Neuropathological and transcriptomic characteristics of the aged brain. Elife 6, e31126 (2017).
Kukull, W. A. et al. Dementia and Alzheimer disease incidence: a prospective cohort study. Arch. Neurol. 59, 1737–1746 (2002).
Larson, E. B. et al. Exercise is associated with reduced risk for incident dementia among persons 65 years of age and older. Ann. Intern. Med. 144, 73–81 (2006).
Crane, P. K. et al. Glucose levels and risk of dementia. N. Engl. J. Med. 369, 540–548 (2013).
Sonnen, J. A. et al. Neuropathology in the adult changes in thought study: a review. J. Alzheimers Dis. 18, 703–711 (2009).
Monoranu, C. M. et al. pH measurement as quality control on human post mortem brain tissue: a study of the BrainNet Europe consortium. Neuropathol. Appl. Neurobiol. 35, 329–337 (2009).
Mills, J. D. et al. The alternative splicing of the apolipoprotein E gene is unperturbed in the brains of Alzheimer’s disease patients. Mol. Biol. Rep. 41, 6365–6376 (2014).
Southworth, L. K., Owen, A. B. & Kim, S. K. Aging mice show a decreasing correlation of gene expression within genetic modules. PLoS Genet. 5, e1000776 (2009).
Levy, O. et al. Age-related loss of gene-to-gene transcriptional coordination among single cells. Nat. Metab. 2, 1305–1315 (2020).
Fujie, S. et al. Aerobic exercise restores aging-associated reductions in arterial adropin levels and improves adropin-induced nitric oxide-dependent vasorelaxation. J. Am. Heart Assoc. 10, e020641 (2021).
Fujie, S. et al. Association between aerobic exercise training effects of serum adropin level, arterial stiffness, and adiposity in obese elderly adults. Appl. Physiol. Nutr. Metab. 42, 8–14 (2017).
Yang, C., DeMars, K. M., Hawkins, K. E. & Candelario-Jalil, E. Adropin reduces paracellular permeability of rat brain endothelial cells exposed to ischemia-like conditions. Peptides 81, 29–37 (2016).
Imaninezhad, M. et al. Directed and enhanced neurite outgrowth following exogenous electrical stimulation on carbon nanotube-hydrogel composites. J. Neural Eng. 15, 056034 (2018).
Meijering, E. et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. A 58, 167–176 (2004).
Pemberton, K., Mersman, B. & Xu, F. Using ImageJ to Assess Neurite Outgrowth in Mammalian Cell Cultures: research Data Quantification Exercises in Undergraduate Neuroscience Lab. J. Undergrad. Neurosci. Educ. 16, A186–A194 (2018).
Clarke, L. E. et al. Normal aging induces A1-like astrocyte reactivity. Proc. Natl Acad. Sci. USA 115, E1896–E1905 (2018).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Yu, L. et al. Adropin preserves the blood-brain barrier through a Notch1/Hes1 pathway after intracerebral hemorrhage in mice. J. Neurochem. 143, 750–760 (2017).
Castanho, I. et al. Transcriptional Signatures of Tau and Amyloid Neuropathology. Cell Rep. 30, 2040–2054 e2045 (2020).
Flurkey, K., Currer, J. M. & Harrison, D. E. in The Mouse in Biomedical Research. 2nd Edn (eds J.G. Fox et al.) (Elsevier, 2007).
Arch, J. R., Hislop, D., Wang, S. J. & Speakman, J. R. Some mathematical and technical issues in the measurement and interpretation of open-circuit indirect calorimetry in small animals. Int J. Obes. (Lond.) 30, 1322–1331 (2006).
Magistretti, P. J. & Allaman, I. A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901 (2015).
Ouimet, B. et al. Motor learning deficits and striatal GSK-3 hyperactivity in Akt3 knockout mice. Behav. Neurosci. 133, 135–143 (2019).
Gomez-Pinilla, F. Brain foods: the effects of nutrients on brain function. Nat. Rev. Neurosci. 9, 568–578 (2008).
St-Onge, M. P. et al. Fasting plasma adropin concentrations correlate with fat consumption in human females. Obes. (Silver Spring) 22, 1056–1063 (2014).
Stevens, J. R. et al. Inverse association between carbohydrate consumption and plasma adropin concentrations in humans. Obes. (Silver Spring) 24, 1731–1740 (2016).
van der Klaauw, A. A. et al. Divergent effects of central melanocortin signalling on fat and sucrose preference in humans. Nat. Commun. 7, 13055 (2016).
Hill, C. M. et al. FGF21 and the physiological regulation of macronutrient preference. Endocrinology 161, bqaa019 (2020).
Kwon, O. S., Andtbacka, R. H. I., Hyngstrom, J. R. & Richardson, R. S. Vasodilatory function in human skeletal muscle feed arteries with advancing age: the role of adropin. J. Physiol. 597, 1791–1804 (2019).
Fouda, A. Y., Fagan, S. C. & Ergul, A. Brain Vasculature and Cognition. Arterioscler Thromb. Vasc. Biol. 39, 593–602 (2019).
Abondio, P. et al. The genetic variability of APOE in different human populations and its implications for longevity. Genes 10, 222 (2019).
Mei, H. et al. Expression array annotation using the BioMediator biological data integration system and the BioConductor analytic platform. AMIA Annu. Symp. Proc. 2003, 445–449 (2003).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Subramanian, A., Kuehn, H., Gould, J., Tamayo, P. & Mesirov, J. P. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23, 3251–3253 (2007).
Getz, A. M., Xu, F., Visser, F., Persson, R. & Syed, N. I. Tumor suppressor menin is required for subunit-specific nAChR alpha5 transcription and nAChR-dependent presynaptic facilitation in cultured mouse hippocampal neurons. Sci. Rep. 7, 1768 (2017).
Mersman, B., Zaidi, W., Syed, N. I. & Xu, F. Taurine Promotes Neurite Outgrowth and Synapse Development of Both Vertebrate and Invertebrate Central Neurons. Front. Synaptic Neurosci. 12, 29 (2020).
Pemberton, K., Mersman, B. & Xu, F. Using ImageJ to Assess Neurite Outgrowth in Mammalian Cell Cultures: Research Data Quantification Exercises in Undergraduate Neuroscience Lab. J. Undergrad. Neurosci. Educ. 16, A186–A194 (2018).
Hammond, R. S., Tull, L. E. & Stackman, R. W. On the delay-dependent involvement of the hippocampus in object recognition memory. Neurobiol. Learn Mem. 82, 26–34 (2004).
Pedros, I. et al. Early alterations in energy metabolism in the hippocampus of APPswe/PS1dE9 mouse model of Alzheimer’s disease. Biochim. Biophys. Acta 1842, 1556–1566 (2014).
Acknowledgements
A.A.B. acknowledges the financial support provided by Saint Louis University and NIH grant R21NS108138. F.X. acknowledges the financial support of Saint Louis University. E.C.J. acknowledges the support provided by NIH grants NS108138 and NS103094. S.G. acknowledges the support of a PSC-CUNY Award, jointly funded by The Professional Staff Congress and The City University of New York. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. Data used for the analyses described in this paper were obtained from the GTEx Portal on 09/14/20. The authors thank Jingying Zhang and Randall L. Mynatt at the Pennington Biomedical Research Center Transgenic Core for construction of the ENHO-Cre mice. The Transgenic Core is supported by the Clinical Nutrition Research Unit Center Grant 1P30 DK072476 entitled “Nutritional Programming: Environmental and Molecular Interactions” sponsored by the National Institutes of Health through the NIDDK (Eric Ravussin, principal investigator). The authors also thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1TR002345 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH.
Author information
Authors and Affiliations
Contributions
A.A.B. was responsible for data management, development of the transgenic mouse models, conceptual development of the project, funding acquisition, and preparation of the paper. C.G., S.A.F. and A.A.B. conceived the original idea, designed experiments, and interpreted data. A.A.B. and J.Z. analyzed RNA seq data from experiment performed in-house and from open access datasets and interpreted the results. S.B., S.G., C.G. and M.L.N. assisted with animal experiments. S.B., S.G., C.G., M.L.N., A.D.N., K.M.D., C.Y., P.J., B.A.M., W.A.B., K.M.H. and E.C-J. collected data. F.X. designed and F.X. and B.A.H. interpreted primary culture experiments that were performed by P.J. and B.A.H. S.B., S.G. and C.G. are equal contributors to the paper, and are co-first authors. A.B. and S.F. laboratories contributed equally to the project, and are co-corresponding authors. All authors read and approved the paper for content.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests. One technology disclosure related to this work was filed by Saint Louis University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Banerjee, S., Ghoshal, S., Girardet, C. et al. Adropin correlates with aging-related neuropathology in humans and improves cognitive function in aging mice. npj Aging Mech Dis 7, 23 (2021). https://doi.org/10.1038/s41514-021-00076-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41514-021-00076-5
This article is cited by
-
Expression of G protein-coupled receptor GPR19 in normal and neoplastic human tissues
Scientific Reports (2023)
-
Low circulating adropin levels in late-middle aged African Americans with poor cognitive performance
npj Aging (2023)
-
Low circulating adropin concentrations predict increased risk of cognitive decline in community-dwelling older adults
GeroScience (2023)