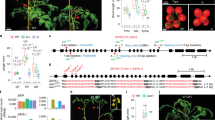
Abstract
Increasing production efficiency is a top priority in agriculture. Optimal plant architecture is the biological basis of dense planting, high crop yield and labour cost savings, and is thus critical for improving agricultural productivity. In cucurbit crops, most species have elongated internodes, but the path to architecture improvement is still not clear. Here we identified a pumpkin accession with a dominant bushy trait, and found that the associated Bush locus harbours a cucurbit-conserved cis-regulatory element in the 5’ untranslated region of a transcription factor gene YABBY1. In cucurbit crops, various B-region deletions enhance the translation of YABBY1, with consequent proportional suppression of stem length in a dose-dependent manner. Depending on different cultivation patterns, the precise deployment of these alleles has significant effects on yield improvement or labour cost saving. Our findings demonstrate that the engineering of the YABBY1 B-region is an efficient strategy to customize plant architecture in cucurbit crops.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Relevant data can be found in the Supplementary Tables. Source data, including raw data for all quantifications, are provided with this paper.
References
Reynolds, M. et al. Addressing research bottlenecks to crop productivity. Trends Plant Sci. 26, 607–630 (2021).
Wang, B., Smith, S. M. & Li, J. Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 437–468 (2018).
Mathan, J., Bhattacharya, J. & Ranjan, A. Enhancing crop yield by optimizing plant developmental features. Development 143, 3283–3294 (2016).
FAOSTAT, Food and Agriculture Organization of the United Nations, https://www.fao.org/home/en (2020).
Loy, J. B. Morpho-physiological aspects of productivity and quality in squash and pumpkins (Cucurbita spp.). Crit. Rev. Plant Sci. 23, 337–363 (2004).
Wehner, T. C. Watermelon. In: Prohens, J., Nuez, F. (eds) Vegetables I: 381–418 (Springer, 2008).
Liu, X., Chen, J. & Zhang, X. Genetic regulation of shoot architecture in cucumber. Hortic. Res. 8, 143 (2021).
Peng, J. et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261 (1999).
Khush, G. S. Green revolution: the way forward. Nat. Rev. Genet. 2, 815–822 (2001).
Wei, C. et al. A point mutation resulting in a 13 bp deletion in the coding sequence of Cldf leads to a GA-deficient dwarf phenotype in watermelon. Hortic. Res. 6, (2019).
Rodríguez-Leal, D. et al. Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480 (2017).
Dogimont, C., Pitrat, M. & McCreight, J. D. 2011 Gene List for Melon Report 33–34 (The Cucurbit Genetics Cooperative, 2011).
Guner, N. & Wehner, T. C. The genes of watermelon. HortScience 39, 1175–1182 (2004).
Ding, W. et al. Fine mapping identified the gibberellin 2-oxidase gene CpDw leading to a dwarf phenotype in squash (Cucurbita pepo L.). Plant Sci. 306, 110857 (2021).
Siegfried, K. R. et al. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128 (1999).
Bowman, J. L. & Smyth, D. R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396 (1999).
Villanueva, J. M. et al. INNER NO OUTER regulates abaxial–adaxial patterning in Arabidopsis ovules. Genes Dev. 13, 3160–3169 (1999).
Eshed, Yuval et al. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131, 2997–3006 (2004).
Golz, J. F., Roccaro, M., Kuzoff, R. & Hudson, A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 131, 3661–3670 (2004).
Juarez, M. T., Twigg, R. W. & Timmermans, M. C. Specification of adaxial cell fate during maize leaf development. Development 131, 4533–4544 (2004).
Jang, S. et al. Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol. Biol. 56, 133–143 (2004).
Yamaguchi, T. et al. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16, 500–509 (2004).
Dai, M. et al. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144, 121–133 (2007).
Kumaran, M. K., Bowman, J. L. & Sundaresan, V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14, 2761–2770 (2002).
Goldshmidt, A., Alvarez, J. P., Bowman, J. L. & Eshed, Y. Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20, 1217–1230 (2008).
Lugassi, N., Nakayama, N., Bochnik, R. & Zik, M. A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis. BMC Plant Biol. 10, 1–13 (2010).
Shi, B. et al. Feedback from lateral organs controls shoot apical meristem growth by modulating auxin transport. Dev. Cell 44, 204–216 (2018).
Zhang, X. L., Yang, Z. P., Zhang, J. & Zhang, L. G. Ectopic expression of BraYAB1-702, a member of YABBY gene family in Chinese cabbage, causes leaf curling, inhibition of development of shoot apical meristem and flowering stage delaying in Arabidopsis thaliana. Int. J. Mol. Sci. 14, 14872–14891 (2013).
Qi, J. et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 45, 1510–1515 (2013).
Sun, H. et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol. Plant 10, 1293–1306 (2017).
Zhao, G. et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 51, 1607–1615 (2019).
Guo, S. et al. Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 51, 1616–1623 (2019).
Leppek, K., Das, R. & Barna, M. Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174 (2018).
Nagai, K. et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 584, 109–114 (2020).
Sarojam, R. et al. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22, 2113–2130 (2010).
Wang, X. et al. Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit. Nat. Plants 7, 419–427 (2021).
Wray, G. A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216 (2007).
Wittkopp, P. J. & Kalay, G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Abe, A. et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174–178 (2012).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Blázquez, M. Quantitative GUS activity assay in intact plant tissue. Cold Spring Harb. Protoc. 2, pdb-prot4688 (2007).
Wang, S. et al. A rare SNP identified a TCP transcription factor essential for tendril development in cucumber. Mol. Plant 8, 1795–1808 (2015).
Xing, H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 1–12 (2014).
Xin, T. et al. Targeted creating new mutants with compact plant architecture using CRISPR/Cas9 genome editing by an optimized genetic transformation procedure in cucurbit plants. Hortic. Res. 9, uhab086 (2022).
Tian, S. et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 36, 399–406 (2017).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Cai, J., Qin, G., Chen, T. & Tian, S. The mode of action of remorin1 in regulating fruit ripening at transcriptional and post‐transcriptional levels. New Phytol. 219, 1406–1420 (2018).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
McCleary, B. V., Solah, V. & Gibson, T. S. Quantitative measurement of total starch in cereal flours and products. J. Cereal Sci. 20, 51–58 (1994).
Paciorek, T., Sauer, M., Balla, J., Wiśniewska, J. & Friml, J. Immunocytochemical technique for protein localization in sections of plant tissues. Nat. Protoc. 1, 104–107 (2006).
Acknowledgements
We thank H. Chen (Hunan Vegetable Research Institute, Hunan Academy of Agricultural Sciences) for providing cucumber seeds used in transformation; J. Wang (Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences) for providing information on watermelon and melon accessions; and X. Zhang (Xinjiang Academy of Agricultural Sciences) for providing melon seeds. This work was supported by the National Natural Science Foundation of China (31701914 to S.W.), the National Key R&D Program of China (2019YFA0906200), the National Natural Science Foundation of China (31922076 to X.Y.), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP) and the Science and Technology Innovation Team of Shaanxi (2021TD-32).
Author information
Authors and Affiliations
Contributions
S.W., X.Y. and S.H. designed the research; K.W., Y.L., J.H., T.X. and H.T. performed genetic transformation; Hongbo Li and S.W. designed the markers; S.W., Y.L., B.W. and K.W. performed vectors construction and most experiments. J.T., G.Z. and Haizhen Li helped to collect seeds; S.W., X.Y., Z.L. and Haizhen Li performed data analysis; S.W. and X.Y. wrote the paper, which was approved by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Qiang Xu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Stem length of Daluo and the bushy SXHD82 accession at 10-week-old stage.
Data are presented as mean ± standard deviation (s.d.) (n = 10 plants). Exact P values are shown (two-tailed, two sample Student’s t-test).
Extended Data Fig. 2 The weight of a single fruit from Daluo and SXHD82 two weeks after pollination.
Data are presented as mean ± SD (n = 12 fruits). Exact P values are shown (two-tailed, two sample Student’s t-test).
Extended Data Fig. 3 CmoCh15G012090 belongs to YABBY transcriptional factor family.
CmoCh15G012090 encodes a protein with a zinc finger-like domain in the N terminus and a YABBY domain in the C terminus.
Extended Data Fig. 4 Quantification of GUS activity of leaves from CmoYABBY1Pro-WT5’ UTR:GUS and CmoYABBY1Pro-Bu5’ UTR:GUS transgenic pumpkin plants.
Data are presented as mean ± SD (n = 4 plants). Exact P values are shown (two-tailed, two sample Student’s t-test).
Extended Data Fig. 5 The specificity of CmoYABBY1 polyclonal antibodies was validated by Western blot analysis.
a, The specific short peptide is selected to design polyclonal antibodies for CmoYABBY1. There are two YABBY1 genes in pumpkin, and CmoCh15G012090 is designated as CmoYABBY1 while CmoCh02G015970 is designated as CmoYABBY1b. The specific short peptide with red underline is selected to design polyclonal antibodies for CmoYABBY1. b, The pCAMBIA 1300-CmoYABBY1-5×myc, 1300-CmoYABBY1b-5×myc, and the corresponding empty vector were transiently expressed in leaves of tobacco (N. benthamiana). Total protein extracted was performed for immunoblotting using the anti-CmoYABBY1 and antic-Myc antibodies. Experiments were repeated independently three times (b).
Extended Data Fig. 6 The protein expression level of CmoYABBY1 detected at different developmental periods of leaves in wildtype pumpkin.
a, The leaves of the three developmental periods from the pumpkin Daluo. Bar = 10 cm. b, The protein expression level of CmoYABBY1 was detected at different developmental periods of leaves in wildtype pumpkin Daluo. Experiments were repeated independently three times (b).
Extended Data Fig. 7 Alignments of YABBY1 5’UTR sequences from species mentioned in Fig.4 (a).
The B-region sequences in the red box are conserved in Cucurbitaceae plants.
Extended Data Fig. 8 Comparison of yield per plant for edited pumpkin.
a, Comparison of yield per plant for WT and edited pumpkin (CmoM-1, CmoM-2, CmoM-3 and CmoM-4) under ample cultivation space conditions. Data are presented as mean ± SD (n = 8 plants). Different letters indicate significant differences (P < 0.05, one-way ANOVA and Tukey’s test). b, The fruits of WT and edited pumpkin (CmoM-1, CmoM-2, CmoM-3 and CmoM-4) under ample cultivation space conditions. Bars = 8 cm.
Extended Data Fig. 9 The editing mutant CmoM-4 enables labor cost saving in greenhouse.
a, WT pumpkin and edited CmoM-4 mutant planted in greenhouse in Beijing, China, in 2021. The CmoM-4 plants showed an upright bushy growth habit without hanging vines, while the WT plants with long and trailing vines require hanging vines and holding fruits. b, Comparison of labor cost time per 100 plants between WT and CmoM-4 in greenhouse (n = 3 biological replicates). Data are presented as mean ± SD. c, Comparison of fruit yield per 100 m2 between WT and CmoM-4 plants in greenhouse (n = 3 biological replicates). Data are presented as mean ± SD. Exact P values are shown (two-tailed, two sample Student’s t-test).
Extended Data Fig. 10 The predicted secondary structures of YABBY1 B-region in cucurbits.
The secondary structures of B-region of CmoYABBY1 (a), CsYABBY1 (b) and ClYABBY1 (c) were predicted by using UNAFold software. (d) Two mutant constructs were used to detect the effect of predicted stem-loop structure on protein translation by determining the LUC/REN activity and mRNA levels. The modified-1 mutant construct was designed to enhance the stem-loop, and the modified-2 mutant construct was designed to destroy the stem-loop. A wild type (WT) construct and those carrying modified-1 or modified-2 constructs were expressed in Cucurbita moschata cotyledons. The mean LUC/REN activity and mRNA levels conferred by each mutant construct were normalized to those of WT control (n = 8 biological replicates). Data are presented as mean ± SD. Different letters indicate significant differences (P < 0.05, one-way ANOVA and Tukey’s test). (e) The secondary structure of mutant B-region (modified-1) of CmoYABBY1 was predicted by UNAFold software.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11.
Supplementary Tables 1–9
Supplementary Table 1 List of pumpkin accessions. Table 2 List of watermelon accessions. Table 3 List of melon accessions. Table 4 Stem length of pumpkin, watermelon and melon accessions. Table 5 Information regarding molecular markers used for fine mapping of Bu. Table 6 Primers used for qPCR. Table 7 Primers used for the construction of native pro::WT/Bu-5’UTR::GUS fusion genes. Table 8 Primers for vector construction of dual-luciferase assay. Table 9 Primers used for CRISPR-Cas9 vector construction in pumpkin, cucumber and watermelon.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data and unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, S., Wang, K., Li, Z. et al. Architecture design of cucurbit crops for enhanced productivity by a natural allele. Nat. Plants 8, 1394–1407 (2022). https://doi.org/10.1038/s41477-022-01297-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01297-6
This article is cited by
-
Accelerating crop domestication through genome editing for sustainable agriculture
Journal of Plant Biochemistry and Biotechnology (2023)
-
Construction and application of a new watermelon germplasm with the phenotype of dwarf and branchless
Functional & Integrative Genomics (2023)
-
CRISPR/Cas9 mediated editing of phytoene desaturase gene in squash
Journal of Plant Biochemistry and Biotechnology (2023)
-
Compact plants enhance productivity
Nature Plants (2022)