Abstract
Gene expression is prone to burst production, making it a highly noisy process that requires additional controls. Upstream open reading frames (uORFs) are widely present in the 5′ leader sequences of 30–50% of eukaryotic messenger RNAs1,2,3. The translation of uORFs can repress the translation efficiency of the downstream main coding sequences. Whether the low translation efficiency leads to a different variation, or noise, in gene expression has not been investigated, nor has the direct biological impact of uORF-repressed translation. Here we show that uORFs achieve low but precise protein production in plant cells, possibly by reducing the protein production rate. We also demonstrate that, by buffering a stable TIMING OF CAB EXPRESSION 1 (TOC1) protein production level, uORFs contribute to the robust operation of the plant circadian clock. Our results provide both an action model and the biological impact of uORFs in translational control to mitigate transcriptional noise for precise protein production.
Similar content being viewed by others
Main
Gene expression is a sequential and highly coordinated process that conveys genetic information in genes (DNA) first to messenger RNAs (mRNAs) and then to functional protein products4. The dynamic and robust gene expression relies on multilayered regulation at the DNA, mRNA and protein levels. Bursts of de novo transcription and translation often result in fluctuations of both mRNA and protein levels and even lead to signal amplifications5. For specific genes, the temporal expression surges may be beneficial5, such as for cell fate determination in Drosophila embryogenesis6 or Arabidopsis sepal morphogenesis7. However, the ‘noisy’ production for most cellular mRNAs may be unproductive, energy consuming or even detrimental to the cells and organisms, especially if the fluctuating mRNAs encode key signalling molecules.
Noise inherited from transcription can be buffered by mRNA degradation8 and translational control. At the level of translation, one effective way to circumvent the negative impact of stochastic mRNA production is to have regulatory information built in cis in mRNAs for translational control. Such information can be the codon usage9, the presence of a secondary structure10 or microRNA target sites in eukaryotic mRNAs11. Another class of cis regulatory sequences is upstream open reading frames (uORFs) present in 5′ leader sequences of 30–50% of mRNAs in diverse organisms1,2,3. The translation of these uORFs can reduce the translation efficiency of the downstream main ORF (mORF) in a broad spectrum of eukaryotes, such as yeast12, human13, vertebrates14 and plants15. The prevalence of uORFs implies their instrumental roles in translation control. These observations motivated us to investigate whether uORFs can mitigate noisy transcription by reducing protein production capacity and variation, and, if so, how do uORFs exert their regulatory role in a biologically relevant context?
Results
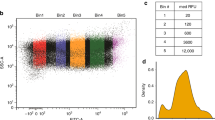
We first examined protein production and variations in single Arabidopsis cells by comparing translation of mORFs in mRNAs with or without uORFs in their 5′ leaders. For an mRNA without uORF(s), ribosomes scan and assemble to translate the mORF (regular translation; Fig. 1a). For an mRNA with translatable uORF(s), translating ribosomes would dissociate after termination or resume scanning for translational re-initiation of the mORF16 (uORF-mediated translation; Fig. 1a). Although the CUG initiation codon is a common alternative initiation codon17,18, only an AUG-initiated, but not CUG-initiated, uORF could impose translation repression of the mORF in plants15 and yeast19. Figure 1b shows combinatorial constructs used to monitor the translation of mORF (enhanced green fluorescent protein (EGFP)) from mRNAs transiently produced in Arabidopsis protoplasts by estradiol induction20, under the control of an AUG-initiated uORF (uORF) or a CUG-initiated uORF (uORFm) in the 5′ leader sequences and with or without the fusion of a protein destabilization signal PEST domain (P)21. mCherry levels were used to infer the transcript levels in flow cytometry assays. We assumed the same transcription rate for EGFP and mCherry under the control of the identical promoters and the positive correlation of mCherry mRNA and protein level.
a, A sketch of regular or uORF-mediated translation via ribosomes (yellow ovals). b, Constructs with dual fluorescence reporters (EGFP and mCherry) under the control of inducible promoters (OLexA), with AUG-initiated or CUG-initiated uORF in the 5′ leader of native EGFP (solid circle, uORF and uORFm). P, PEST domain (solid triangle). c, Background-subtracted mean EGFP levels with s.d. (μ ± σ) across equal-distance mCherry measurement level bins (n = 15–18). Fluorescence was measured by flow cytometry. Three independent experiments were performed with replicates shown in Extended Data Fig. 1. Cell numbers are listed in Source Data Fig. 1. d, The association of mean EGFP signal levels and CV (%CV, σ/μ) in constructs indicated. Error bars represent s.d. (σ) from bootstrapping with 1,000 iterated resampling in d.
In uORFm, the fusion of PEST reduced the EGFP levels produced from transcripts spanning over a magnitude difference in abundance (indicated by mean mCherry level; uORFm, P versus uORFm in Fig. 1c). The reduction in EGFP level is even more evident under the control of uORF (uORF versus uORFm; Fig. 1c and Extended Data Fig. 1a). In addition to the markedly reduced EGFP level, the coefficient of variation (CV, σ/μ) was also much lower for the uORF groups (Fig. 1d). The variations were negatively correlated with EGFP level for both the uORF and uORFm groups (Fig. 1d and Extended Data Fig. 1b), similar to the observation by monitoring the expression variation of 43 yeast proteins under 11 experimental conditions22. The results supported our hypothesis that the uORF-mediated translational repression could effectively attenuate both the protein level and variation inherited from gene expression variabilities in single-cell populations.
Inspired by the dissection of noise reported recently11,23,24,25, we also generated constructs with both EGFP and mCherry under the control of uORF or uORFm to evaluate the impact on both intrinsic and extrinsic noise (Extended Data Fig. 2). The intrinsic noise indicates the amount of variation in one protein (for example, EGFP) that is independent of the other (mCherry), whereas the covariance of the two is essentially referred as the extrinsic noise. In principle, intrinsic noise reflects the variation in the expression process of a gene, and the extrinsic noise refers to the variation among cells, such as cellular resources26,27. However, factors in translation machineries could influence both intrinsic and extrinsic noise in our measurement scheme. The mathematical derivation of intrinsic and extrinsic noise is detailed in the Supplementary Information. The results confirmed that uORFs could effectively repress intrinsic as well as extrinsic noise by translational repression (Extended Data Fig. 2).
We developed a simple mathematical model to better understand the possible mechanism of the reduction in variation of protein levels in single cells. Mass action kinetics were built according to the schemes depicted, including a state of mRNA with uORF occupied with ribosome (mu) in the scenario (Fig. 2). With the linear noise approximation28, a set of mathematical expressions for the variances and covariances of species in the model under the number fluctuation caused by the probabilistic nature of the mass action (biochemical reactions), we fit experimental data for the parameters (detailed in Supplementary Information and Supplementary Table 1).
Illustrations showing reaction species, directions and constants for regular and uORF-mediated translation for modelling. mF and muF, free-form mRNA state without and with uORF, respectively; mu, mRNA state with uORF occupied with ribosome; P and Pu, protein state from regular and uORF-mediated translation, respectively; Reaction rates for the production of mRNA (km) and protein (kp and kpu), the association of ribosome to muF (kon) or dissociation of the ribosome from mu (koff); degradation rates of mF (γmF), muF (γmuF), and mu (γmu)kx, γx.
With the fitted model, the Fano factor (FF) for the uORF-regulated protein can be written as:
where Pu is the mean protein abundance under uORF-mediated translation, Var(Pu) is the corresponding variance, kPu is the protein production rate, γP and γmu are the degradation rates for the protein and mRNA, respectively, and mu denotes the mRNA with uORF occupied with ribosome. In Equation (1), the variation in protein is described by the intrinsic noise (1, the FF for a Poisson distribution) and the noise propagated from the upstream species, mu. Under the limit of higher degradation rate in mu (γmu >> γP) the latter (~kPu/γmu) is approximately the mean amount of protein each mu generates before it degrades, a factor that contributes both the overall protein production and its variation. In the fitted parameters (Supplementary Table 1), the protein production rate is reduced by a factor of >6 (kP/kPu) in the uORF regulation, whereas the degradation rate of mu remains similar to that of regular mRNA (γmu ≈ γmF) detailed in Supplementary Fig. 2, thus indicating that the lower variation of uORF-regulated translation may result from reduced protein production rate. Therefore, by reducing the translational rates, uORFs act to reduce the uncertainty in the protein production.
Protein variations derived from stochastic gene expression affect steady-state proteins but also those functioning in key cellular pathways, such as cell cycle regulation29 and gene regulatory networks30. The oscillating nature of circadian genes also makes them more vulnerable to stochastic gene expression. To put the regulatory roles of uORFs in the biological context, we examined how uORFs contribute to the robust operation of the circadian clock in Arabidopsis.
Organisms on Earth have evolved to possess internal circadian clocks oscillating in ~24-hour cycles to synchronize with the external day/night cycles. The dyssynchronization of internal and external clocks often leads to disease or disadvantageous surviving fitness31,32. TIMING OF CAB EXPRESSION 1 (TOC1) is a key central oscillator of the Arabidopsis circadian clock33,34. Plants with increased TOC1 protein level have circadian period length >24 hours35 and slower cell cycle during vegetative development36, so an appropriate TOC1 protein level may be a prerequisite of optimal growth fitness.
TOC1 mRNA has four AUG-initiated uORFs in its 5′ leader sequence (Extended Data Fig. 3a). Ribosome protection fragments were identified for all four uORFs in a previously reported dataset15, thus suggesting active translation of these uORFs. We then assayed how the translation of these uORFs affects TOC1 protein production and the circadian robustness in plants. Mutations of AUGs to stop codons (TAG or TGA) (uORFm–TOC1; Extended Data Fig. 3b) led to increased level and variation of the reporter protein EGFP in Arabidopsis protoplasts (Extended Data Fig. 3c,d), which confirms that these uORFs have the potential to effectively buffer TOC1 protein production.
We next examined whether uORFs in TOC1 mRNA contribute to the translational control of TOC1 protein production in plants by complementing the toc1-101 mutant with a TOC1 mini-gene35 carrying the wild-type TOC1 5′ leader with uORFs or with each AUG mutated to a stop codon (uORF–TOC1 and uORFm–TOC1, respectively; Fig. 3a). Independent transgenic lines with transgene mRNA levels spanning an order of magnitude (Extended Data Fig. 4) were selected to investigate the inter-relationship of TOC1 mRNA, TOC1 protein and circadian period length (inferred by the pCCA1:LUC2 reporter gene) (Fig. 3a).
a, Schematic illustration of constructs used to complement toc1-101 mutant carrying the pCCA1:LUC2 reporter and experimental design for measuring RNA/protein levels at ZT14 and circadian period lengths as described in Methods. b,c, Scatter dot plots showing correlations of TOC1 RNA and TOC1–YFP protein levels (b) and circadian period length (c) in independent uORF–TOC1 and uORFm–TOC1 transgenic lines (n = 11). TOC1 RNA and protein levels are in arbitrary units (a.u.). One-way statistical analysis of covariance (ANCOVA) for two independent groups are provided in Source Data Fig. 3. Regression analysis indicated with P values and R2 values. Three independent experiments were performed and are shown Extended Data Fig. 7.
At comparable mRNA levels, increased TOC1-yellow fluorescent protein (YFP) protein levels were produced from the transgene with uORFm–TOC1 compared with uORF–TOC1 (Fig. 3b and Extended Data Figs. 4, 5 and 7a). Therefore, uORFm–TOC1 transgenic lines acquire significantly longer circadian periods than the uORF–TOC1 lines (Fig. 3c and Extended Data Figs. 6 and 7b). These results suggest that uORFs function to confine the TOC1 protein level, which is crucial for plants to sustain a 24-hour circadian clock.
The clock behaviour in a whole plant is a combined result of cell-to-cell, even organ-to-organ, communication37,38. We next examined whether uORFs also exert regulatory roles in TOC1 protein production and clock precision at the level of the individual cells. A time-course quantification of nucleus-localized TOC1–YFP fusion protein in cells isolated from seven independent lines was conducted (four for uORF–TOC1 and three for uORFm–TOC1) (Extended Data Fig. 8). The results showed more synchronized circadian oscillation of TOC1–YFP in individual cells from uORF–TOC1 lines than in uORFm–TOC1 cells with comparable TOC1–YFP levels (Fig. 4a,b and Extended Data Figs. 8, 9 and 10a). The peak phases are less coherent and period lengths broadly distributed for the uORFm–TOC1 cells (Fig. 4c,d and Extended Data Fig. 10b,c). These data supported that, without functional uORFs in the 5′ leader, TOC1–YFP oscillations are much noisier among cells, thus compromising the clock robustness in individual cells.
a, Heat maps showing time course (hours in continuous dark, DD) of mean TOC1–YFP level in nuclei of cells isolated from representative uORF–TOC1 and uORFm–TOC1 lines. Horizontal lines are individual cells ranked by k-means clustering. Scaled fluorescence intensity from minimum to maximum shown in the pseudo-colour bar. b, Mean ± s.e.m. of TOC1–YFP protein levels through time. Individual lines marked in different colours with number (n) of cells analysed. Black line, toc1-101 negative cells. c, Violin plots of curve-fitted peak phase distribution of individual cells from each line through time (hours). d, Violin plots of period length distribution defined by successive curve-fitted peaks. Both the first and/or the second period length for each line were plotted. Letters denote statistically significant subgroups (two-tailed F-test, P < 0.001). Three independent experiments were performed and are shown in Extended Data Figs. 9 and 10. Data used are listed in Source Data Fig. 4.
In the Arabidopsis circadian clock, TOC1 and CCA1 represent evening and morning genes, respectively, and mutually inhibit the expression of each other, thus constituting a double negative feedback loop34,39. Variable TOC1 expression phases and periods cannot ensure the expression precision of the morning gene CCA1, which tips the balance of the Arabidopsis circadian clock. These results together support that uORF-repressed translation is required for plant cells to maintain an optimal and precise TOC1 protein level to sustain a robust circadian clock.
Conclusions
In this Letter, we demonstrate that uORFs can effectively reduce both the intrinsic and extrinsic noise from stochastic gene expression (Extended Data Fig. 2). Protein production and variation are both attenuated by uORFs in individual cells expressing a wide range of mRNA levels (reporter constructs in Fig. 1 and the endogenous plant gene TOC1 in Fig. 4). This is primarily achieved by the reduced protein production rate (Supplementary Table 1) in uORF-mediated translation. We also show that uORFs are vital for TOC1 protein accumulation with precision to sustain a robust clock in Arabidopsis (Figs. 3c and 4). Although circadian clocks in plants and animals both operate via feedback loops, they harbour distinct core clock components. Analogous to TOC1 in the plant clock, the translation of some core clock genes in mice (Arntl, Clock, Cry1, Nr1d1 and Nr1d2) was previously found to be regulated by uORFs40. Whether uORFs also contribute to the clock robustness in animals is worth investigating.
uORFs have been implicated to function in diverse physiological processes in eukaryotes, such as metabolite homoeostasis in plants41,42,43 and the association with human disease44,45. uORFs are also frequently observed in genes encoding protein kinases and transcription factors3,46. Studying whether uORFs function to ensure stable and precise signalling cascades potentiated by these signalling molecules would be of great interest.
The homoeostasis of a physiological response in a multicellular organism could be achieved by cell–cell coupling47. To maintain protein homoeostasis within a single cell, the use of uORFs in translation control demands a relatively lower energy budget, such as the passive reduction of noise by cellular compartmentation48, compared with routes via mRNA and/or protein degradation or using integral feedbacks that often require extrinsic biomolecules49,50. Hence, uORFs represent one significant mode of translational control, functioning cooperatively with additional regulatory mechanisms to ensure an effective and meticulous protein translation with low energy expenditure.
Methods
Constructs for protoplast transient assays
For dual fluorescence reporter assays (Fig. 1), DNA fragments harbouring LexA–VP16–ER (XVE)-inducible LexA operon–35S promoter and TNos–OLexA–MCS–T3A were PCR amplified from pER10–Bar51 and used to substitute the 35S promoter and Nos terminator in p326 35S:smGFP vector to generate p326 OLexA–smGFP–TNos–OLexA–MCS–T3A. smGFP was later replaced by EGFP from pCAMBIA1390 35S:FD-MCP-EGFP. The mCherry coding sequence from p326 35S:mCherry was subcloned into MCS to create p326 OLexA–EGFP–ZmSP–TNos–OLexA–mCherry–ZmSP–T3A. The maize SPERMINE SYNTHASE 1 PEST sequence (ZmSP)21 was fused in-frame with EGFP or mCherry as a protein destabilization signal. DNA fragments harbouring the 5′ leader sequence of AT3G55520 (AtFKBP20-1) carrying an AUG-uORF or a site-directed mutagenized CUG-uORF were inserted into the 5′ end of EGFP for single uORF/uORFm constructs (Fig. 1 and Extended Data Fig. 1) and into both the 5′ ends of EGFP and mCherry for dual uORF/uORFm constructs (Extended Data Fig. 2).
For translation activity quantification of uORFs in AT5G61380 (TOC1) (Extended Data Fig. 3), the wild-type TOC1 5′ leader (uORF–TOC1) was amplified from genomic DNA. The four AUG-uORFs in TOC1 5′ leader were point mutated into stop codons (ATG131→TAG, ATG255→TGA, ATG329→TAG, and ATG464→TAG) by overlapping PCR and termed uORFm–TOC1. uORF–TOC1 and uORFm–TOC1 were subcloned separately into the 5′ end of EGFP in p326 OLexA–EGFP–ZmSP–TNos–OLexA–mCherry–ZmSP–T3A. All primers used are listed in Supplementary Table 2.
Dual fluorescence assays in Arabidopsis protoplasts
Transgenic plants expressing XVE transcription activator52 were established by transforming wild-type Arabidopsis thaliana (Col-0) with pER10-Bar vector51 with Agrobacterium tumefaciens strain GV3101. Successful expression of XVE protein was confirmed by immunoblotting with antiserum against LexA (Abcam, ab14553,1:10,000 dilution). Rosette leaves of 4-week-old transgenic plants grown under 12-hour light/dark at 22 °C were used for protoplast preparation as described53 by adopting a sandwich method54 and the use of W5 medium with 5 mM glucose for protoplast suspension. Each transfection involved 3.0 × 105 protoplasts and 60 μg of plasmid DNA prepared by using Plasmid Maxi kits (QIAGEN). p326 35S:EGFP and p326 35S:mCherry were included as controls. XVE was induced to express in protoplasts by 2 μM β-estradiol for 16–40 hours. The quantification of EGFP (excitation 488 nm, emission 530/30 nm), mCherry (excitation 561 nm, emission 620/15 nm) and chlorophyll autofluorescence (excitation 488 nm, emission 695/40 nm) was accomplished by using the Attune NxT Flow Cytometer and software version 2.7 and version 3.1 (Thermo Fisher Scientific) and FlowJo version 10.
mCherry levels were classified by equal-distance binning in log-space for mean mCherry levels. The mean EGFP levels (μ) and standard deviation (σ) were then calculated for each mCherry bin after background subtraction. The 1,000 bootstrapping iterations of 50% subsampling were used to evaluate the standard deviation of the coefficient of variation (σ/μ) sampled.
For RNA levels and half-life measurements of uORF/uORFm-regulated EGFP transcripts (Supplementary Figs. 2 and 4), transfected protoplasts were induced to express the transgene by 16-hour β-estradiol treatment, followed by treatment with 10 μM actinomycin D for 1, 2 and 4 hours before cells were collected and used for RNA preparation. The NucleoSpin RNA XS column (Macherey-Nagel) was used for RNA isolation, followed by reverse transcription with SuperScript IV reverse transcriptase (Thermo Fisher Scientific) and quantitative PCR (qPCR) as described55. Primers used in this study are in Supplementary Table 2.
Generation and characterization of uORF–TOC1 and uORF m –TOC1 transgenic plants
A DNA fragment harbouring an uORFm–TOC1 leader sequence was used to replace the uORF–TOC1 leader sequence in pZP221 TMG–YFP35 to create pZP221 TMG (uORFm)–YFP. Primers used to generate uORFm–TOC1 are in Supplementary Table 2. TMG (uORF)–YFP and TMG (uORFm)–YFP in binary vectors were introduced separately into the toc1-101 mutant carrying the pCCA1:LUC2 reporter gene56 by Agrobacterium GV3101.
For protein and RNA analyses (Fig. 3b,c and Extended Data Figs. 4, 5, 7 and 8), 12-day-old seedlings from T2 generations of TMG (uORF)–YFP and TMG (uORFm)–YFP transgenic seedlings grown on 1/2 Murashige and Skoog medium (MS) with gentamycin under 16-hour light/8-hour dark conditions were collected at Zeitgeber time 14 (ZT14). Total RNA was extracted with the pine-tree method57 and quantified by qPCR as described55 with QuantStudio 12K Flex software (version 1.2.2 for data acquisition and version 1.4 for data analysis) except for the primers used for TOC1 in Supplementary Information Table 2. Total protein was extracted in freshly prepared extraction buffer58 (100 mM Tris-HCl, pH 7.8, 4 M urea, 5% SDS, 15% glycerol, protease inhibitor cocktail and 5% β-mercaptoethanol) and separated with NuPAGE Bis-Tris 4–12% gel. TOC1–YFP protein was detected by immunoblot analyses with anti-GFP antisera (GenLab, catalogue number 50005-05, 1:10,000 dilution) by Amersham Hyperfilm ECL (GE Healthcare) for SuperSignal (Thermo Fisher Scientific) linear chemiluminescence signal detection. Coomassie Brilliant Blue (Sigma-Aldrich)-stained membranes were loading controls. Data were analysed by using the Fiji_win64 image processing package59 (Extended Data Fig. 5). For Extended Data Fig. 8, immunoblot quantitation was performed by using UVP ChemStudio Plus Touch (Analytik Jena) for capturing linear chemiluminescence signals, and VisionWorks v9.1.20063.7760 was used for data acquisition and analysis.
For calculating the circadian period length (Fig. 3c and Extended Data Fig. 7), 7-day-old seedlings were transferred to 96-well black plates with 1/2 MS medium containing 0.5 mM luciferin (PerkinElmer). A light-proof chamber (Phenotron, HiPoint) with a SOPHIA 2048BX Large Format CCD Camera (Princeton Instruments) with LightField v6.10 was used to measure bioluminescence under constant light condition for 4 days (LL; 24–120 hours). A custom-made software, ImagePro v6.2, was used to acquire data from images for fast Fourier transform-nonlinear least squares (FFT-NLLS) analysis with the Biological Rhythms Analysis Software System (BRASS v3.0)60.
For measuring TOC1–YFP levels in single cells (Fig. 4 and Extended Data Figs. 9 and 10), TMG (uORF)–YFP and TMG (uORFm)–YFP transgenic plants were grown on 1/2 MS with gentamycin under 16-hour light/8-hour dark and 22 °C. Rosette leaves of 4-week-old plants were used to prepare protoplasts as described above. Protoplasts were recovered in W5 medium for 48 hours under an entrainment condition, transferred to darkness and imaged by Leica Application Suite X_4.1.0.23081 using STELLARIS 8 (Leica) confocal microscopy for YFP signals (excitation 488 nm, emission 518–581 nm) hourly at times corresponding to ZT12 to ZT80. Image analyses and quantification of TOC1–YFP signals in nuclei are described in ‘Analysis for single-cell trajectories’ in Supplementary Information. The raw data were used for generating heat maps, and averaged oscillation curves are shown in Fig. 4a,b and Extended Data Figs. 9 and 10a. Curved fitted data used for peak identification and period length calculation and violin plots were prepared with ggplot2 package version 3.3.5 in R x64 4.0.3 in Fig. 4c,d and Extended Data Fig. 10b,c.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Source data are provided with this paper.
Code availability
All codes used in this study are available at https://github.com/cherrihsu/uORF_dataprocess.
References
Dever, T. E., Ivanov, I. P. & Sachs, M. S. Conserved upstream open reading frame nascent peptides that control translation. Annu. Rev. Genet. 54, 237–264 (2020).
Andrews, S. J. & Rothnagel, J. A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15, 193–204 (2014).
von Arnim, A. G., Jia, Q. & Vaughn, J. N. Regulation of plant translation by upstream open reading frames. Plant Sci. 214, 1–12 (2014).
Crick, F. Central dogma of molecular biology. Nature 227, 561–563 (1970).
Eldar, A. & Elowitz, M. B. Functional roles for noise in genetic circuits. Nature 467, 167–173 (2010).
Bothma, J. P. et al. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc. Natl Acad. Sci. USA 111, 10598–10603 (2014).
Roeder, A. H. K. Use it or average it: stochasticity in plant development. Curr. Opin. Plant Biol. 41, 8–15 (2018).
Baudrimont, A., Jaquet, V., Wallerich, S., Voegeli, S. & Becskei, A. Contribution of RNA degradation to intrinsic and extrinsic noise in gene expression. Cell Rep. 26, 3752–3761 (2019).
Blake, W. J., Kaern, M., Cantor, C. R. & Collins, J. J. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003).
Zydowicz-Machtel, P., Swiatkowska, A., Popenda, L., Gorska, A. & Ciesiolka, J. Variants of the 5ʹ-terminal region of p53 mRNA influence the ribosomal scanning and translation efficiency. Sci. Rep. 8, 1533 (2018).
Schmiedel, J. M. et al. MicroRNA control of protein expression noise. Science 348, 128–132 (2015).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Calvo, S. E., Pagliarini, D. J. & Mootha, V. K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl Acad. Sci. USA 106, 7507–7512 (2009).
Chew, G. L., Pauli, A. & Schier, A. F. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun. 7, 11663 (2016).
Liu, M. J. et al. Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25, 3699–3710 (2013).
Hinnebusch, A. G., Ivanov, I. P. & Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 (2016).
Lee, S. et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl Acad. Sci. USA 109, E2424–E2432 (2012).
Ingolia, N. T., Lareau, L. F. & Weissman, J. S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 (2011).
Brar, G. A. et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 (2012).
Zuo, J. R., Niu, Q. W. & Chua, N. H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 (2000).
Maruri-Lopez, I. et al. A maize spermine synthase 1 PEST sequence fused to the GUS reporter protein facilitates proteolytic degradation. Plant Physiol. Biochem. 78, 80–87 (2014).
Bar-Even, A. et al. Noise in protein expression scales with natural protein abundance. Nat. Genet. 38, 636–643 (2006).
Araujo, I. S. et al. Stochastic gene expression in Arabidopsis thaliana. Nat. Commun. 8, 2132 (2017).
Alamos, S., Reimer, A., Niyogi, K. K. & Garcia, H. G. Quantitative imaging of RNA polymerase II activity in plants reveals the single-cell basis of tissue-wide transcriptional dynamics. Nat. Plants https://doi.org/10.1038/s41477-021-00976-0 (2021).
Hani, S. et al. Live single-cell transcriptional dynamics via RNA labelling during the phosphate response in plants. Nat. Plants https://doi.org/10.1038/s41477-021-00981-3 (2021).
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002).
Swain, P. S., Elowitz, M. B. & Siggia, E. D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12795–12800 (2002).
Elf, J. & Ehrenberg, M. Fast evaluation of fluctuations in biochemical networks with the linear noise approximation. Genome Res. 13, 2475–2484 (2003).
Novak, B. & Tyson, J. J. Mechanisms of signalling–memory governing progression through the eukaryotic cell cycle. Curr. Opin. Cell Biol. 69, 7–16 (2021).
Nagata, S. & Kikuchi, M. Emergence of cooperative bistability and robustness of gene regulatory networks. PLoS Comput. Biol. 16, e1007969 (2020).
Dodd, A. N. et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633 (2005).
Bass, J. & Lazar, M. A. Circadian time signatures of fitness and disease. Science 354, 994–999 (2016).
Mas, P., Alabadi, D., Yanovsky, M. J., Oyama, T. & Kay, S. A. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236 (2003).
Pokhilko, A., Mas, P. & Millar, A. J. Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 7, 23 (2013).
Mas, P., Kim, W. Y., Somers, D. E. & Kay, S. A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570 (2003).
Fung-Uceda, J. et al. The circadian clock sets the time of DNA replication licensing to regulate growth in Arabidopsis. Dev. Cell 45, 101–113 (2018).
Endo, M., Shimizu, H., Nohales, M. A., Araki, T. & Kay, S. A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515, 419–422 (2014).
Takahashi, N., Hirata, Y., Aihara, K. & Mas, P. A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell 163, 148–159 (2015).
Alabadi, D. et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883 (2001).
Janich, P., Arpat, A. B., Castelo-Szekely, V., Lopes, M. & Gatfield, D. Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res. 25, 1848–1859 (2015).
Rahmani, F. et al. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 150, 1356–1367 (2009).
Laing, W. A. et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27, 772–786 (2015).
van der Horst, S., Filipovska, T., Hanson, J. & Smeekens, S. C. M. Metabolite control of translation by conserved peptide uORFs: the ribosome as a metabolite multi-sensor. Plant Physiol. 182, 110–122 (2020).
Lee, D. S. M. et al. Disrupting upstream translation in mRNAs is associated with human disease. Nat. Commun. 12, 1515 (2021).
Schulz, J. et al. Loss-of-function uORF mutations in human malignancies. Sci. Rep. 8, 2395 (2018).
Ye, Y. H. et al. Analysis of human upstream open reading frames and impact on gene expression. Hum. Genet. 134, 605–612 (2015).
Smith, S. & Grima, R. Single-cell variability in multicellular organisms. Nat. Commun. 9, 345 (2018).
Stoeger, T., Battich, N. & Pelkmans, L. Passive noise filtering by cellular compartmentalization. Cell 164, 1151–1161 (2016).
Briat, C., Gupta, A. & Khammash, M. Antithetic integral feedback ensures robust perfect adaptation in noisy biomolecular networks. Cell Syst. 2, 15–26 (2016).
Aoki, S. K. et al. A universal biomolecular integral feedback controller for robust perfect adaptation. Nature 570, 533–537 (2019).
Tsai, H. L. et al. HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell 26, 2858–2872 (2014).
Jianru Zuo, P. D. H. & Chua, Nam-Hai Applications of chemical-inducible expression systems in functional genomics and biotechnology. Methods Mol. Biol. 323, 329–342 (2006).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Wu, F. H. et al. Tape–Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009).
Wu, J. F. et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 7, 13181 (2016).
Wang, Y. et al. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23, 486–498 (2011).
Chang, S., Puryear, J. & Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116 (1993).
Chang, C. S. J., Maloof, J. N. & Wu, S. H. COP1-Mediated Degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156, 228–239 (2011).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Southern, M. M., Brown, P. E. & Hall, A. in Arabidopsis Protocols (eds Salinas, J. & Sanchez-Serreno, J. J.) 293–305 (Humana Press, Totowa, NJ, 2006).
Acknowledgements
We thank members of the Wu and Hsu labs, T. Oyama and H.-L. Tsai for discussion; Y. Wang for comments on the manuscript and the illustrations; the assistance of M.-J. Fang, J.-Y. Huang for image capturing and analyses with Leica Stellaris 8 at the Cell Biology Core, Institute of Plant and Microbial Biology (IPMB), Academia Sinica; the assistance of T.-W. Tai for analysis of flow cytometry at the flow cytometry core facility of the Institute of Biomedical Sciences, Academia Sinica; the technical support of A. Tseng at the IPMB Flow Cytometry Analysis and Sorting Service, Academia Sinica; the assistance of S.-J. Chou, A.-P. Chen, M.-J. Fang and M.-L. Cheng for RNA and DNA analysis at the Genomic Technology Core, IPMB, Academia Sinica; and the help of C.-W. Wang, T.-S. Yu, H.-H. Lin and P. Mas for providing experimental materials or reagents. Funding: this work was supported by research grants to S.-H.W. from the Ministry of Science and Technology, Taiwan (107-2321-B-001-007-, 108-2321-B-001-023-, 109-2811-B-001-595- and 110-2326-B-001-018-), research grants to C.-P.H. from the Ministry of Science and Technology, Taiwan (109-2113-M-001-022-MY4 and 110-2123-M-001-005-), the investigator award from Academia Sinica (AS-IA-106-M01) and intramural grants from the Institute of Plant and Microbial Biology and the Institute of Chemistry, Academia Sinica, Taiwan.
Author information
Authors and Affiliations
Contributions
H.-W.W., C.-P.H. and S.-H.W. conceived and designed the experiments and wrote the manuscript. H.-W.W. and J.-F.W. performed the experiments and analysed the data. H.-W.W., E.F., C.-C.S.Y. and C.-P.H. conducted the modelling and analysed the data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Christine Queitsch, Simon Alamos and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Replicate results showing that AUG-initiated uORF attenuates EGFP signal levels and noise.
Background subtracted mean EGFP levels with SD (μ±𝜎) across equal-distance mCherry bins (a), and the coefficient of variation (%CV) vs mean EGFP levels (μ) (b) in constructs shown in Fig. 1c, d. Error bars represent standard deviations (𝜎) from bootstrapping with 1,000 iterated resampling in b. Results shown are replicates 2 and 3 of independent experiments from dual fluorescence assays (Fig. 1). Cell numbers are listed in Source Data_Fig. 1).
Extended Data Fig. 2 uORF-mediated translation represses mean signal level and variation in dual uORF/uORFm constructs.
a, Constructs with dual uORF/uORFm in both fluorescence reporters (EGFP and mCherry) under the control of inducible promoters (OLexA), with AUG- or CUG-initiated uORF in the 5' leader of native EGFP and mCherry. b, Mean EGFP signal levels with SD (μ±𝜎) across equal-distance bins (n = 4-8) of mCherry+EGFP signal levels after background subtraction. Fluorescence was measured by flow cytometry. Three independent experiments performed. Cell numbers are listed (Source Data ED_Fig. 2). c, The association of mean EGFP signal levels and coefficient of variation (%CV, 𝜎/μ) in constructs indicated. Error bars represent standard deviations (𝜎) from bootstrapping with 1,000 iterated resampling. Error bars smaller than dots were not shown. d, The quantification of EGFP signal noise in CV square (●) from extrinsic (■) and intrinsic noise (◆) in dual fluorescence constructs expressed in cell populations for three independent experiments. Error bars represent standard deviations (𝜎) from bootstrapping with 1,000 iterated resampling.
Extended Data Fig. 3 Triplicate results showing that uORFs of TOC1 attenuate mean EGFP signals and variation in Arabidopsis protoplasts.
a, Schematic illustration of the TOC1 5’ leader sequence carrying 4 AUG-initiated uORFs. b, Constructs with dual fluorescence reporters (EGFP and mCherry) under the control of inducible promoters (OLexA), with uORFs initiated with AUG or stop codons in the 5’ leader of EGFP (uORF-TOC1 and uORFm-TOC1, respectively). P: PEST domain. c, Mean EGFP levels with SD (μ±𝜎) across equal-distance mCherry bins (n = 14-15) after background subtraction. Fluorescence was measured by flow cytometry. Cell numbers are listed (Source Data_ED_Fig. 3). d, The association of mean EGFP levels and coefficient of variation (%CV, 𝜎/μ) in constructs indicated. Error bars represent standard deviations (𝜎) from bootstrapping with 1,000 iterated resampling.
Extended Data Fig. 4 Triplicate results for TOC1-YFP mRNA levels in transgenic lines used.
Scatter dot plot showing TOC1-YFP mRNA levels at ZT14 in 12-d-old T2 seedlings of uORF-TOC1 and uORFm-TOC1 lines used in this study (bulked n = 16-20). Dots and error bars show mean RNA values ±SD of three technical replicates with three independent quantifications. Results were normalized to UBQ10 mRNA. Wild-type (Col-0) and toc1-101-carrying pCCA1:LUC2 plants were included for comparison.
Extended Data Fig. 5 Triplicate results for TOC1-YFP protein levels in transgenic lines used.
a, TOC1-YFP protein levels at ZT14 in 12-d-old T2 seedlings of uORF-TOC1 and uORFm-TOC1 lines used in this study visualized and quantified by immunoblotting with anti-GFP antisera. *: non-specific signals. Results from three independent quantifications are shown. b, Scatter dot plot showing TOC1-YFP protein levels normalized to Coomassie Blue-stained gels (boxed areas with dashed white lines in a). Wild-type (Col-0) and toc1-101-carrying pCCA1:LUC2 plants were included for comparison.
Extended Data Fig. 6 Triplicate results for period lengths in transgenic lines used.
Scatter dot plots showing circadian period lengths of individual uORF-TOC1 and uORFm-TOC1 lines used in this study. Results were calculated from LUC2 data under continuous light (LL; 24-120 h). Three independent quantifications were performed. Dots and error bars show mean period length ±SD from a population of seedlings (n = 13-16 with RAE < 0.3) with three independent quantifications. Wild-type (Col-0) and toc1-101-carrying pCCA1:LUC2 plants were included for comparison.
Extended Data Fig. 7 Replicate results showing that uORFs attenuate TOC1 protein production to sustain a 24-h circadian clock.
Scatter dot plots showing correlations of TOC1 RNA with TOC1-YFP protein levels (a), with circadian period lengths (b) in replicates 2 and 3 shown in Fig. 3b, c, and TOC1-YFP protein levels with circadian period lengths (c) in 3 biological replicates in uORF-TOC1 and uORFm-TOC1 lines (n = 11). Statistical analysis of covariance, one-way ANCOVA for 2 independent groups, and regression analysis indicated with p-values and R2 values.
Extended Data Fig. 8 Triplicate results showing TOC1-YFP RNA and protein levels in representative lines used for single cell experiments in Fig. 4.
a, TOC1-YFP mRNA levels at ZT14 in 12-d-old T2 seedlings of uORF-TOC1 and uORFm-TOC1 lines used in this study. Data and error bars show as mean RNA values ±SD of three technical replicates with three independent quantifications. Results were normalized to the level of UBQ10 mRNA. b, TOC1-YFP protein levels at ZT14 in 12-d-old T2 transgenic seedlings of uORF-TOC1 and uORFm-TOC1 lines indicated were visualized and quantified by immunoblot with anti-GFP antisera. *: non-specific signals. Numbers indicated TOC1-YFP protein levels normalized to Coomassie Blue-stained gels (boxed areas with dashed white lines) with TOC1-YFP level in Te3 set to 1. toc1-101-carrying pCCA1:LUC2 plants was included for comparison. Results from 3 biological replicates are shown.
Extended Data Fig. 9 Replicate results showing that cellular rhythms are more robust in uORFs cells.
Results are replicates 2 and 3 of single cell time-course experiments shown in Fig. 4a. Heatmaps showing time-course (h in continuous dark, DD) mean TOC1-YFP level in nuclei of cells isolated from representative uORF-TOC1 and uORFm-TOC1 lines. Horizontal lines are individual cells ranked by k-means clustering. Scaled fluorescence intensity from minimum to maximum shown in the pseudo-colour bar. Data used were listed in Source Data Fig. 4.
Extended Data Fig. 10 Replicate results showing that uORFs confine TOC1-YFP protein expression phase and period length.
Results are replicates 2 and 3 of single cell time-course experiments shown in Fig. 4b, c, d. a, Mean TOC1-YFP protein levels through time. Error bar as standard error of measurements. b, Violin plots of curve-fitted peak phase distribution of individual cells from each line through time (h). c, Violin plots of period length distribution defined by successive curve-fitted peaks. Letters denote statistically significant subgroups (two-tailed F-test, p < 0.001). Both the first and/or the second period length for each line were plotted. Letters denote statistically significant subgroups (two-tailed F-test, p < 0.001). Data used were listed in Source Data Fig. 4.
Supplementary information
Supplementary Information
Supplementary Discussion, Figs. 1–6, and Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1 and Extended Data Fig. 1.
Source Data Fig. 3
Statistical source data for Fig. 3 and Extended Data Fig. 7.
Source Data Fig. 4
Statistical source data for Fig. 4, and Extended Data Figs. 9 and 10.
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Figs. 4–6.
Source Data Extended Data Fig. 5
Unprocessed blots and gels for Extended Data Fig.5a.
Source Data Extended Data Fig. 8
Statistical source data for Extended Data Fig. 8.
Source Data Extended Data Fig. 8
Unprocessed blots and gels for Extended Data Fig. 8b.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, HW., Fajiculay, E., Wu, JF. et al. Noise reduction by upstream open reading frames. Nat. Plants 8, 474–480 (2022). https://doi.org/10.1038/s41477-022-01136-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-022-01136-8
This article is cited by
-
ProTInSeq: transposon insertion tracking by ultra-deep DNA sequencing to identify translated large and small ORFs
Nature Communications (2024)
-
A natural uORF variant confers phosphorus acquisition diversity in soybean
Nature Communications (2022)
-
Noise-cancelling translation syncs cellular clocks
Nature Plants (2022)