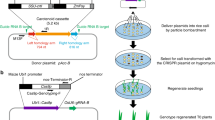
Abstract
Structural variations (SVs), such as inversion and duplication, contribute to important agronomic traits in crops1. Pan-genome studies revealed that SVs were a crucial and ubiquitous force driving genetic diversification2,3,4. Although genome editing can effectively create SVs in plants and animals5,6,7,8, the potential of designed SVs in breeding has been overlooked. Here, we show that new genes and traits can be created in rice by designed large-scale genomic inversion or duplication using CRISPR/Cas9. A 911 kb inversion on chromosome 1 resulted in a designed promoter swap between CP12 and PPO1, and a 338 kb duplication between HPPD and Ubiquitin2 on chromosome 2 created a novel gene cassette at the joint, promoterUbiquitin2::HPPD. Since the original CP12 and Ubiquitin2 genes were highly expressed in leaves, the expression of PPO1 and HPPD in edited plants with homozygous SV alleles was increased by tens of folds and conferred sufficient herbicide resistance in field trials without adverse effects on other important agronomic traits. CRISPR/Cas-based genome editing for gene knock-ups has been generally considered very difficult without inserting donor DNA as regulatory elements. Our study challenges this notion by providing a donor-DNA-free strategy, thus greatly expanding the utility of CRISPR/Cas in plant and animal improvements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study have been deposited into the NCBI Sequence Read Archive under accession number PRJNA765115 and CNGB Sequence Archive (CNSA)40 of China National GeneBank DataBase (CNGBdb)41 with accession number CNP0001922. Source data are provided with this paper.
References
Tao, Y., Zhao, X., Mace, E., Henry, R. & Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 12, 156–169 (2019).
Liu, Y. et al. Pan-genome of wild and cultivated soybeans. Cell 182, 162–176 (2020).
Alonge, M. et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145–161 (2020).
Jayakodi, M. et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284–289 (2020).
Schwartz, C. et al. CRISPR-Cas9-mediated 75.5-Mb inversion in maize. Nat. Plants 6, 1427–1431 (2020).
Korablev, A. N., Serova, I. A. & Serov, O. L. Generation of megabase-scale deletions, inversions and duplications involving the Contactin-6 gene in mice by CRISPR/Cas9 technology. BMC Genet. 18, 112 (2017).
Schmidt, C. et al. Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. Nat. Commun. 11, 4418 (2020).
Beying, N., Schmidt, C., Pacher, M., Houben, A. & Puchta, H. CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 6, 638–645 (2020).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A. & Voytas, D. F. DNA replicons for plant genome engineering. Plant Cell 26, 151–163 (2014).
Dahan-Meir, T. et al. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95, 5–16 (2018).
Lu, Y. et al. Targeted, efficient sequence insertion and replacement in rice. Nat. Biotechnol. 38, 1402–1407 (2020).
Qin, P. et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184, 3542–3558 (2021).
Li, J. et al. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J. Mol. Cell. Biol. 7, 284–298 (2015).
Qi, Y. et al. Targeted deletion and inversion of tandemly arrayed genes in Arabidopsis thaliana using zinc finger nucleases. G3 3, 1707–1715 (2013).
Siehl, D. L. et al. Broad 4-hydroxyphenylpyruvate dioxygenase inhibitor herbicide tolerance in soybean with an optimized enzyme and expression cassette. Plant Physiol. 166, 1162–1176 (2014).
Wang, H. et al. Bipyrazone: a new HPPD-inhibiting herbicide in wheat. Sci. Rep. 10, 5521 (2020).
Lermontova, I. & Grimm, B. Overexpression of plastidic protoporphyrinogen IX oxidase leads to resistance to the diphenyl-ether herbicide acifluorfen. Plant Physiol. 122, 75–84 (2000).
Xing, H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Wang, J. & Oard, J. H. Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep. 22, 129–134 (2003).
Shen, R. et al. Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice. Nat. Commun. 8, 1310 (2017).
Wang, Y. et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948 (2015).
Zhang, J. et al. Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc. Natl Acad. Sci. USA 113, E5163–E5171 (2016).
Guan, J. et al. Genome structure variation analyses of peach reveal population dynamics and a 1.67 Mb causal inversion for fruit shape. Genome Biol. 22, 13 (2021).
Li, Y. et al. A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 196, 282–291 (2012).
Zhang, H. et al. Gain-of-function of the 1-aminocyclopropane-1-carboxylate synthase gene ACS1G induces female flower development in cucumber gynoecy. Plant Cell 33, 306–321 (2021).
Schmidt, C., Pacher, M. & Puchta, H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 98, 577–589 (2019).
Xiang, G. et al. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell. Mol. Life Sci. 75, 4619–4628 (2018).
Xie, Y. et al. The intronic cis element SE1 recruits trans-acting repressor complexes to repress the expression of Elongated Uppermost Internode1 in rice. Mol. Plant 11, 720–735 (2018).
Zhang, H. et al. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898 (2018).
Shi, J. et al. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216 (2017).
Shan, Q., Wang, Y., Li, J. & Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410 (2014).
Ding, J. et al. Validation of a rice specific gene, sucrose phosphate synthase, used as the endogenous reference gene for qualitative and real-time quantitative PCR detection of transgenes. J. Agric. Food Chem. 52, 3372–3377 (2004).
Chen, Y. et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 7, 1–6 (2018).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Henry, I. M., Dilkes, B. P., Miller, E. S., Burkart-Waco, D. & Comai, L. Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics 186, 1231–1245 (2010).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Marcais, G. et al. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14, e1005944 (2018).
Guo, X. Q. et al. CNSA: a data repository for archiving omics data. Database 2020, baaa055 (2020).
Chen, F. Z. et al. CNGBdb: China national geneBank database. Hereditas 42, 799–809 (2020).
Acknowledgements
This work was supported by funds from Qingdao Kingagroot Compounds Company and grants from National Natural Science Foundation of China (award no. 31872933 to L.J.).
Author information
Authors and Affiliations
Contributions
L.J., H.L., J.-K.Z. and B.S. supervised this project. Y. Lu, J.W., B.C., S.M. and L.L. performed or led key experiments with the help from other authors: Q.C., Yong Li and D.D. for genotyping; Q. Hou and G.L. for protein expression and purification; Yucai Li, Y.D., J.W., Q. Hu, T.C., Y. Luo., Y.W. and Z.L. for vector construction and rice transformation; and C.S., Y.Z. and G.C. for field tests. S.S. and G.F. performed genome SV analysis using NGS deep sequencing data. J.-K.Z., H.L. and L.J. wrote the manuscript with contributions from other co-authors. L.J. conceived the idea.
Corresponding authors
Ethics declarations
Competing interests
A patent was filed to patent office in 2019 in China and L.J., H.L., J.W., Y. Lu and S.M. were listed as inventors. All other authors have no competing interests.
Additional information
Peer review information Nature Plants thanks Yaoguang Liu, Sergei Svitashev and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Tables 1–5.
Source data
Source Data Fig. 1
Uncropped and unprocessed scans of gels for Fig. 1c.
Source Data Fig. 3
Uncropped and unprocessed scans of gels for Fig. 3d.
Rights and permissions
About this article
Cite this article
Lu, Y., Wang, J., Chen, B. et al. A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat. Plants 7, 1445–1452 (2021). https://doi.org/10.1038/s41477-021-01019-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-01019-4
This article is cited by
-
Hidden prevalence of deletion-inversion bi-alleles in CRISPR-mediated deletions of tandemly arrayed genes in plants
Nature Communications (2023)
-
Regulation of gene-edited plants in Europe: from the valley of tears into the shining sun?
aBIOTECH (2023)
-
Trait Improvement of Solanaceae Fruit Crops for Vertical Farming by Genome Editing
Journal of Plant Biology (2023)
-
Genome Editing Technology and Its Application to Metabolic Engineering in Rice
Rice (2022)
-
Massive crossover suppression by CRISPR–Cas-mediated plant chromosome engineering
Nature Plants (2022)