Abstract
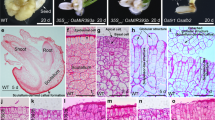
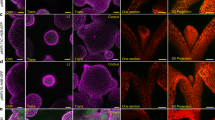
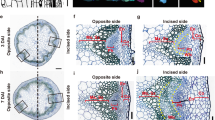
In plant tissue culture, callus forms from detached explants in response to a high-auxin-to-low-cytokinin ratio on callus-inducing medium. Callus is a group of pluripotent cells because it can regenerate either roots or shoots in response to a low level of auxin on root-inducing medium or a high-cytokinin-to-low-auxin ratio on shoot-inducing medium, respectively1. However, our knowledge of the mechanism of pluripotency acquisition during callus formation is limited. On the basis of analyses at the single-cell level, we show that the tissue structure of Arabidopsis thaliana callus on callus-inducing medium is similar to that of the root primordium or root apical meristem, and the middle cell layer with quiescent centre-like transcriptional identity exhibits the ability to regenerate organs. In the middle cell layer, WUSCHEL-RELATED HOMEOBOX5 (WOX5) directly interacts with PLETHORA1 and 2 to promote TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 expression for endogenous auxin production. WOX5 also interacts with the B-type ARABIDOPSIS RESPONSE REGULATOR12 (ARR12) and represses A-type ARRs to break the negative feedback loop in cytokinin signalling. Overall, the promotion of auxin production and the enhancement of cytokinin sensitivity are both required for pluripotency acquisition in the middle cell layer of callus for organ regeneration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The sequence data can be accessed at the Arabidopsis Genome Initiative (https://www.arabidopsis.org/) under the following accession numbers: WOX11 (AT3G03660), WOX5 (AT3G11260), WOX7 (AT5G05770), PLT1 (AT3G20840), PLT2 (AT1G51190), SCR (AT3G54220), ARR5 (AT3G48100), ARR7 (AT1G19050), ARR12 (AT2G25180), ARR2 (AT4G16110), WUS (AT2G17950), JKD (AT5G03150), TAA1 (AT1G70560), TAR2 (AT4G24670), KCS6 (AT1G68530), PDF1 (AT2G42840), ATML1 (AT4G21750), BDG1 (AT1G64670), PIN2 (AT5G57090), SCZ (AT1G46264), NPY4 (AT2G23050), HAN (AT3G50870), SMXL3 (AT3G52490), TMO5 (AT3G25710), WAT1 (AT1G75500), ANT (AT4G37750), ATHB8 (AT4G32880), 4CL1 (AT1G51680), PXY (AT5G61480), bHLH068 (AT4G29100), TAN1 (AT3G05330) and HIK (AT1G18370). The RNA-seq and single-cell RNA-seq data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession numbers GSE156990, GSE178354 and GSE156991. The RNA-seq and single-cell RNA-seq data can be accessed using the online tool (http://xulinlab.cemps.ac.cn/), and gene IDs can be used to search for gene expression patterns. The data that support the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.
References
Ikeuchi, M. et al. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406 (2019).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Sugimoto, K., Jiao, Y. & Meyerowitz, E. M. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471 (2010).
Trinh, D.-C. et al. PUCHI regulates very long chain fatty acid biosynthesis during lateral root and callus formation. Proc. Natl Acad. Sci. USA 116, 14325–14330 (2019).
Abe, M., Takahashi, T. & Komeda, Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 26, 487–494 (2001).
Kurdyukov, S. et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18, 321–339 (2006).
Sessions, A., Weigel, D. & Yanofsky, M. F. The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263 (1999).
Benková, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003).
Di Laurenzio, L. et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433 (1996).
ten Hove, C. A. et al. SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr. Biol. 20, 452–457 (2010).
Welch, D. et al. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21, 2196–2204 (2007).
Aida, M. et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 (2004).
Sarkar, A. K. et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814 (2007).
Efroni, I., Ip, P.-L., Nawy, T., Mello, A. & Birnbaum, K. D. Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 16, 9 (2015).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Brady, S. M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007).
De Rybel, B. et al. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437 (2013).
Cheng, Y., Qin, G., Dai, X. & Zhao, Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl Acad. Sci. USA 105, 21017–21022 (2008).
Nawy, T. et al. The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell 19, 103–113 (2010).
Wallner, E.-S. et al. Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr. Biol. 27, 1241–1247 (2017).
Smetana, O. et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 565, 485–489 (2019).
Baima, S. et al. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121, 4171–4182 (1995).
Etchells, J. P. & Turner, S. R. The PXY–CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137, 767–774 (2010).
Lee, D., Ellard, M., Wanner, L. A., Davis, K. R. & Douglas, C. J. The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol. Biol. 28, 871–884 (1995).
Ranocha, P. et al. WALLS ARE THIN 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63, 469–483 (2010).
Zhang, Y. et al. Two types of bHLH transcription factor determine the competence of the pericycle for lateral root initiation. Nat. Plants 7, 633–643 (2021).
Liu, J. et al. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093 (2014).
Tanaka, H. et al. The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 9, 1199–1211 (2004).
Walker, K. L., Müller, S., Moss, D., Ehrhardt, D. W. & Smith, L. G. Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17, 1827–1836 (2007).
Gordon, S. P. et al. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134, 3539–3548 (2007).
Kurihara, D., Mizuta, Y., Sato, Y. & Higashiyama, T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015).
Zhai, N. & Xu, L. CRE/LOX-based analysis of cell lineage during root formation and regeneration in Arabidopsis. aBIOTECH 1, 153–156 (2020).
Burkart, R. C. et al. PLETHORA and WOX5 interaction and subnuclear localisation regulates Arabidopsis root stem cell maintenance. Preprint at bioRxiv https://doi.org/10.1101/818187 (2019).
Kareem, A. et al. PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25, 1017–1030 (2015).
Kim, J.-Y. et al. Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J. 37, e98726 (2018).
Santuari, L. et al. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28, 2937–2951 (2016).
Kieber, J. J. & Schaller, G. E. Cytokinins. Arabidopsis Book 12, e0168 (2014).
To, J. P. C. et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671 (2004).
Liu, Z. et al. The type-B cytokinin response regulator ARR1 inhibits shoot regeneration in an ARR12-dependent manner in Arabidopsis. Plant Cell 32, 2271–2291 (2020).
Dai, X. et al. ARR12 promotes de novo shoot regeneration in Arabidopsis thaliana via activation of WUSCHEL expression. J. Integr. Plant Biol. 59, 747–758 (2017).
Meng, W. J. et al. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29, 1357–1372 (2017).
Zhang, T.-Q. et al. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29, 1073–1087 (2017).
Matosevich, R. et al. Local auxin biosynthesis is required for root regeneration after wounding. Nat. Plants 6, 1020–1030 (2020).
Matosevich, R. & Efroni, I. The quiescent center and root regeneration. J. Exp. Bot. https://doi.org/10.1093/jxb/erab319 (2021).
Cheng, Z. J. et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161, 240–251 (2013).
Radhakrishnan, D. et al. A coherent feed forward loop drives vascular regeneration in damaged aerial organs growing in normal developmental-context. Development 147, dev185710 (2020).
Leibfried, A. et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175 (2005).
Ishihara, H. et al. Primed histone demethylation regulates shoot regenerative competency. Nat. Commun. 10, 1786 (2019).
Yang, W., Choi, M.-H., Noh, B. & Noh, Y.-S. De novo shoot regeneration controlled by HEN1 and TCP3/4 in Arabidopsis. Plant Cell Physiol. 61, 1600–1613 (2020).
Ikeuchi, M., Sugimoto, K. & Iwase, A. Plant callus: mechanisms of induction and repression. Plant Cell 25, 3159–3173 (2013).
Hu, X. & Xu, L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172, 2363–2373 (2016).
Kanei, M., Horiguchi, G. & Tsukaya, H. Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development 139, 2436–2446 (2012).
Huang, X. et al. The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Mol. Plant 11, 970–982 (2018).
Stepanova, A. N. et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008).
Zürcher, E. et al. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161, 1066–1075 (2013).
Wang, Q. et al. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nat. Commun. 11, 679 (2020).
He, C., Chen, X., Huang, H. & Xu, L. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 8, e1002911 (2012).
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K. & Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 (2005).
Taniguchi, M., Sasaki, N., Tsuge, T., Aoyama, T. & Oka, A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 48, 263–277 (2007).
Wu, F. H. et al. Tape–Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009).
Shimotohno, A., Heidstra, R., Blilou, I. & Scheres, B. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 32, 1085–1100 (2018).
Zhang, G. et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 5, 491–497 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Haga, N. et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 157, 706–717 (2011).
Kobayashi, K. et al. Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J. 34, 1992–2007 (2015).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Acknowledgements
We thank ABRC, T. Xu and J.-W. Wang for providing mutant seeds and marker lines. This work was supported by grants from the National Natural Science Foundation of China (no. 31630007), the Strategic Priority Research Program of the Chinese Academy of Sciences (no. XDB27030103), Youth Innovation Promotion Association CAS (no. 2014241) and the National Key Laboratory of Plant Molecular Genetics to L.X.
Author information
Authors and Affiliations
Contributions
N.Z. and L.X. designed the research, analysed the data and wrote the article. N.Z. performed the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Kalika Prasad, Pil Joon Seo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, legend of Video 1, legends of Tables 1–7 and references.
Supplementary Video 1
WOX5 expression pattern in callus on CIM.
Supplementary Tables
Supplementary Table 1. List of cell clusters 0 to 9. Supplementary Table 2. GO analysis of cell cluster 2. Supplementary Table 3. List of genes in RNA-seq analysis of Col-0 and wox5-1 wox7-1 calli. Supplementary Table 4. List of genes in RNA-seq analysis of Col-0 and plt1-21 plt2-21 calli. Supplementary Table 5. List of genes regulated by WOX5/7 and PLT1/2. Supplementary Table 6. List of primers used in this study. Supplementary Table 7. List of genes related to the cell cycle.
Source data
Source Data Fig. 2
Unprocessed western blots for Fig.2b.
Source Data Fig. 3
Unprocessed western blots for Fig.3e.
Rights and permissions
About this article
Cite this article
Zhai, N., Xu, L. Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants 7, 1453–1460 (2021). https://doi.org/10.1038/s41477-021-01015-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-01015-8
This article is cited by
-
Coordination between two cis-elements of WRKY33, bound by the same transcription factor, confers humid adaption in Arabidopsis thaliana
Plant Molecular Biology (2024)
-
Transcriptome analysis reveals the effect of propyl gallate on kiwifruit callus formation
Plant Cell Reports (2024)
-
Single-cell resolution analysis reveals the preparation for reprogramming the fate of stem cell niche in cotton lateral meristem
Genome Biology (2023)
-
WOX11: the founder of plant organ regeneration
Cell Regeneration (2023)
-
Application of single-cell multi-omics approaches in horticulture research
Molecular Horticulture (2023)