Abstract
The assembly of light-harvesting chlorophyll-binding proteins (LHCPs) is coordinated with chlorophyll biosynthesis during chloroplast development. The ATP-independent chaperone known as chloroplast signal recognition particle 43 (cpSRP43) mediates post-translational LHCP targeting to the thylakoid membrane and also participates in tetrapyrrole biosynthesis (TBS). How these distinct actions of cpSRP43 are controlled has remained unclear. Here, we demonstrate that cpSRP43 effectively protects several TBS proteins from heat-induced aggregation and enhances their stability during leaf greening and heat shock. While the substrate-binding domain of cpSRP43 is sufficient for chaperoning LHCPs, the stabilization of TBS clients requires the chromodomain 2 of the protein. Strikingly, cpSRP54—which activates cpSRP43’s LHCP-targeted function—inhibits the chaperone activity of cpSRP43 towards TBS proteins. High temperature weakens the interaction of cpSRP54 with cpSRP43, thus freeing cpSRP43 to interact with and protect the integrity of TBS proteins. Our data indicate that the temperature sensitivity of the cpSRP43–cpSRP54 complex enables cpSRP43 to serve as an autonomous chaperone for the thermoprotection of TBS proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data supporting the findings of this study are available within the article and its Supplementary Information. Source data are provided with this paper.
References
Mochizuki, N. et al. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15, 488–498 (2010).
Jarvis, P. & Lopez-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802 (2013).
Allen, J. F., de Paula, W. B., Puthiyaveetil, S. & Nield, J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 16, 645–655 (2011).
Plumley, G. F. & Schmidt, G. W. Light-harvesting chlorophyll a/b complexes: interdependent pigment synthesis and protein assembly. Plant Cell 7, 689–704 (1995).
Dall’Osto, L., Bressan, M. & Bassi, R. Biogenesis of light harvesting proteins. Biochim. Biophys. Acta 1847, 861–871 (2015).
Wang, P. & Grimm, B. Connecting chlorophyll metabolism with accumulation of the photosynthetic apparatus. Trends Plant Sci. https://doi.org/10.1016/j.tplants.2020.12.005 (2021).
Wang, P. & Grimm, B. Organization of chlorophyll biosynthesis and insertion of chlorophyll into the chlorophyll-binding proteins in chloroplasts. Photosynth. Res. https://doi.org/10.1007/s11120-015-0154-5 (2015).
Brzezowski, P., Richter, A. S. & Grimm, B. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 1847, 968–985, (2015).
Tanaka, R. & Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 (2007).
Masuda, T. Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth. Res. 96, 121–143 (2008).
Adams, N. B. P. et al. The active site of magnesium chelatase. Nat. Plants 6, 1491–1502 (2020).
Larkin, R. M., Alonso, J. M., Ecker, J. R. & Chory, J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906 (2003).
Peter, E. & Grimm, B. GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol. Plant 2, 1198–1210 (2009).
Richter, A. S. et al. Phosphorylation of GENOMES UNCOUPLED 4 alters stimulation of Mg chelatase activity in angiosperms. Plant Physiol. 172, 1578–1595 (2016).
Tanaka, R., Kobayashi, K. & Masuda, T. Tetrapyrrole metabolism in Arabidopsis thaliana. Arabidopsis Book 9, e0145 (2011).
Zhu, J. K. Abiotic stress signaling and responses in plants. Cell 167, 313–324 (2016).
Chen, B., Retzlaff, M., Roos, T. & Frydman, J. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 3, a004374 (2011).
Sangwan, V., Orvar, B. L., Beyerly, J., Hirt, H. & Dhindsa, R. S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 31, 629–638 (2002).
Wang, Q. L., Chen, J. H., He, N. Y. & Guo, F. Q. Metabolic reprogramming in chloroplasts under heat stress in plants. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19030849 (2018).
Hu, S., Ding, Y. & Zhu, C. Sensitivity and responses of chloroplasts to heat stress in plants. Front Plant Sci. 11, 375 (2020).
Ha, J. H., Lee, H. J., Jung, J. H. & Park, C. M. Thermo-induced maintenance of photo-oxidoreductases underlies plant autotrophic development. Dev. Cell 41, 170–179 e174 (2017).
Ziehe, D., Dünschede, B. & Schünemann, D. Molecular mechanism of SRP-dependent light-harvesting protein transport to the thylakoid membrane in plants. Photosynth. Res. 138, 303–313 (2018).
Akopian, D., Shen, K., Zhang, X. & Shan, S. O. Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721 (2013).
Schünemann, D. et al. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc. Natl Acad. Sci. USA 95, 10312–10316 (1998).
Li, X., Henry, R., Yuan, J., Cline, K. & Hoffman, N. E. A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc. Natl Acad. Sci. USA 92, 3789–3793 (1995).
Jaru-Ampornpan, P. et al. ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat. Struct. Mol. Biol. 17, 696–702 (2010).
Liang, F. C. et al. Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proc. Natl Acad. Sci. USA 113, E1615–E1624 (2016).
Siegel, A. et al. A disorder-to-order transition activates an ATP-independent membrane protein chaperone. J. Mol. Biol. 432, 166708 (2020).
Jaru-Ampornpan, P., Chandrasekar, S. & Shan, S. O. Efficient interaction between two GTPases allows the chloroplast SRP pathway to bypass the requirement for an SRP RNA. Mol. Biol. Cell 18, 2636–2645 (2007).
Chandrasekar, S., Sweredoski, M. J., Sohn, C. H., Hess, S. & Shan, S. O. Co-evolution of two GTPases enables efficient protein targeting in an RNA-less chloroplast signal recognition particle pathway. J. Biol. Chem. 292, 386–396 (2017).
Moore, M., Harrison, M. S., Peterson, E. C. & Henry, R. Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275, 1529–1532 (2000).
Wang, P. et al. Chloroplast SRP43 acts as a chaperone for glutamyl-tRNA reductase, the rate-limiting enzyme in tetrapyrrole biosynthesis. Proc. Natl Acad. Sci. USA 115, E3588–E3596 (2018).
Klimyuk, V. I. et al. A chromodomain protein encoded by the Arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11, 87–99 (1999).
Amin, P. et al. Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 121, 61–70 (1999).
Jonas-Straube, E., Hutin, C., Hoffman, N. E. & Schunemann, D. Functional analysis of the protein-interacting domains of chloroplast SRP43. J. Biol. Chem. 276, 24654–24660 (2001).
Dunschede, B., Bals, T., Funke, S. & Schunemann, D. Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J. Biol. Chem. 286, 35187–35195 (2011).
Falk, S. & Sinning, I. The C terminus of Alb3 interacts with the chromodomains 2 and 3 of cpSRP43. J. Biol. Chem. 285, le25–le26 (2010).
Holdermann, I. et al. Chromodomains read the arginine code of post-translational targeting. Nat. Struct. Mol. Biol. 19, 260–263 (2012).
Wang, P., Richter, A. S., Kleeberg, J. R. W., Geimer, S. & Grimm, B. Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat. Commun. 11, 1254 (2020).
Apel, K. & Hirt, H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 (2004).
Herbst, J., Hey, D. & Grimm, B. Posttranslational control of tetrapyrrole biosynthesis: interacting proteins, chaperones, auxiliary factors. Adv. Bot. Res. 91, 163–194 (2019).
Kotak, S. et al. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316 (2007).
Onate-Sanchez, L. & Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1, 93 (2008).
Zhou, S., Sawicki, A., Willows, R. D. & Luo, M. C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett. 586, 205–210 (2012).
Jaru-Ampornpan, P. et al. Mechanism of an ATP-independent protein disaggregase: II. distinct molecular interactions drive multiple steps during aggregate disassembly. J. Biol. Chem. 288, 13431–13445 (2013).
Acknowledgements
We thank D. Schünemann (Ruhr-Universität Bochum) for providing us the chaos and ffc mutants, the GST–cpSRP43 expression vector, and antibodies against cpSRP43 and cpSRP54, and P. Hardy for critical reading of the manuscript. This work was supported by a grant from the Chinese Scholarship Council to S.J., grant nos R35 GM136321 and DOE.DE-SC0020661 to A.S. and S.S., and grants from the Deutsche Forschungsgemeinschaft to B.G. (nos FOR2092, GR 936/18-1 and SFB TRR175, subproject C04) and to P.W. (no. WA 4599/2-1).
Author information
Authors and Affiliations
Contributions
B.G. and P.W. designed the experiments. S.J. and A.S. performed the experiments. S.J., A.S., S.S., B.G. and P.W. analysed the data. B.G. and P.W. wrote the manuscript. S.J., A.S. and S.S. gave critical comments and revisions to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Plants thanks Briardo Llorente, Tatsuru Masuda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 cpSRP54 deficiency does not compromise the abundance of GluTR, CHLH, and GUN4.
a, Phenotype of 18-day-old cpsrp54 knock-out mutant (ffc) and wild-type (Col-0) in long-day (18 h light/ 6 h dark). Scale bar, 1 cm. b, Steady-state levels of TBS proteins in the wild-type and ffc were detected by immunoblotting using the indicated antibodies. Ponceau S stained large subunit of RubisCo (RbcL) and Actin are shown as loading controls. c, Semiquantitative analysis with Image J software (NIH) of the immunoblots in (b). The relative amounts of GluTR, CHLH and GUN4 in ffc were normalized to the levels in Col-0. The data are plotted as means ± s.d. (n = 3 independent biological replicates). The statistical analyses were performed using two-tailed Student’s t-tests. No asterisk indicates no significant differences compared to protein levels in Ler-0: * P < 0.05, **P < 0.01.
Extended Data Fig. 2 Physical interaction of cpSRP43 with CHLH and GUN4.
a, Immunoblotting analyses of protein extracts after the BiFC assay (shown in Fig. 2a) confirmed expression of cpSRP43-YFPn, GUN4-YFPn (c-Myc antibody) and YFPc tagged CHLM (GFP antibody) and CHLH (CHLH antibody). b, Immunoblotting analyses confirmed expression of cpSRP43-YFPn and GUN4-YFPc shown in Fig. 2a. Three biological repeats have been performed with similar results.
Extended Data Fig. 3 In vitro enzymatic assays of MgCh.
a, Time course curves of the MgCh enzymatic assay with fluorescence detection of MgP (Ex 416 nm/Em 595 nm). The data are plotted as mean ± s.d. (n = 3 independent experiments). b, Recombinant proteins used in the assay were stained by Coomassie Brilliant Blue.
Extended Data Fig. 4 Short-term heat stress does not alter the transcriptional levels of GluTR, CHLH, GUN4, cpSRP43 and cpSRP54.
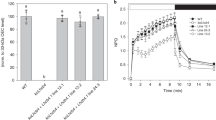
Relative mRNA levels of indicated genes in 18-day-old Ler-0, chaos and cpSRP43-OX were measured prior to and after 2 h heat treatment at 42 °C. Gene expression was calculated relative to the Ler-0 and SAND was used as the reference gene. N.D., not detected. The data are plotted as mean ± s.d. (n = 3 independent biological repeats). The small open circles represent the individual values. Letters above histograms indicate significant differences as determined by two-way ANOVA with Tukey test (P < 0.05).
Extended Data Fig. 5 Measurement of the binding affinity of cpSRP43 with GUN4 using microscale thermophoresis (MST).
The fluorophore-labelled GUN4 were used at constant concentration (20 nM), while the concentration of the non-labelled wild-type (a) or truncated cpSRP43 (b-e) were varied between 3.05 nM and 100 μM. N.D., not detectable. The data are plotted as mean ± s.d. (n = 3 independent experiments).
Extended Data Fig. 6 Accumulation of Chl and Chl precursors in various chaos complementation lines.
HPLC analyses of Chl (a), Proto, MgP and MgPMME (b) and Pchlide (c) in the seedlings of 18-day-old Ler, chaos and a set of chaos complementation lines. FW, fresh weight. The data are plotted as mean ± s.d. (n = 3 independent biological repeats), the small open circles represent the individual values. Letters above histograms indicate significant differences as determined by one-way ANOVA with Tukey test (P < 0.05).
Extended Data Fig. 7 The cpSRP54M peptide stimulates the chaperone activity of cpSRP43 cysless towards LHCP.
The turbidity of 1 µM LHCP diluted out of 8 M urea was measured in the presence of cysteine-mutated cpSRP43 (cpSRP43 C175A/C297S) with (red line) or without (blue line) 50 μM cpSRP54M peptide present. The turbidity at 360 nm was normalized to the value at 0 μM cpSRP43, and the lines are fits of the data to Eq. 1 in the Methods, and the obtained Ksol values were 2 ± 0.4 μM and 0.03 ± 0.09 μM for apo cpSRP43 and cpSRP43 plus cpSRP54M peptide, respectively. This experiment was performed for one time, the similar results have been obtained in the previous publication27.
Extended Data Fig. 8 Comparison of cpSRP43 chaperone activity towards GUN4 and GluTR in the presence of cpSRP54M peptide, cpSRP54M domain, and full-length mature cpSRP54.
Heat-induced aggregation of GluTR (a) and GUN4 (b) was monitored in the presence of apo cpSRP43 (blue lines in each panel) and compared to cpSRP43 with 50 μM cpSRP54M peptide (red lines in the left panels), 20 μM cpSRP54M domain (red lines in the middle panels), or 10 μM full length (FL) mature cpSRP54 (red lines in the right panels). Additional experiments were carried out with cpSRP43 and each cpSRP54 variant in the absence of either client (grey lines in each panel). The turbidity at 360 nm was normalized to the value at 0 μM cpSRP43 under each condition, plotted as mean ± s.e. (n = 3 independent experiments), and fit to Eq. 1 in the Methods. Data and Ksol values for apo cpSRP43 are the same as in Figs. 3a and c. Ksol values for cpSRP43 in the presence of cpSRP54M variants were not obtained, as no protection was observed.
Extended Data Fig. 9 cpSRP54 deficiency does not compromise Chl biosynthesis during heat treatment.
HPLC analyses of Chl (a), MgP and MgPMME (b) and Pchlide (c) in 18-day-old Col-0 and ffc prior to and after 2 or 6 h heat treatment at 42 °C. DW, dry weight. The data are plotted as mean ± s.d. (n = 3 independent biological repeats). The small open circles represent the individual values. Letters above histograms indicate significant differences as determined by two-way ANOVA with Tukey test (P< 0.05).
Extended Data Fig. 10 Heat treatment promotes the binding affinity of cpSRP43 with GluTR, CHLH and GUN4.
a, Co-immunoprecipitation assay to determine the changes in the interaction of cpSRP43 with GluTR, CHLH and GUN4 prior to or after heat treatment at 42 °C for 2 hours (h). Total chloroplast extracts from wild type (Ler-0, used as negative control) and transgenic Arabidopsis lines overexpressing cpSRP43-FLAG were incubated with anti-FLAG affinity beads. cpSRP43 interaction partners were detected by immunoblotting analyses using the indicated antibodies. b-d, In vitro His pull-down assay showing the enhanced binding affinity of GST-cpSRP43 with His-tagged GluTR (b), CHLH (c) and GUN4 (d) at high temperature. Recombinant purified His-tagged GluTR, CHLH and GUN4 (5 μM) were used as bait and incubated with GST or GST-cpSRP43 (5 μM) at 15 °C or 30 °C for 1 h. The proteins bound to His-tagged GluTR, CHLH and GUN4 were eluted with the elution buffer containing 250 mM imidazole. Input and elution fractions were analysed by immunoblot analyses using the indicated antibodies. Two independent biological repeats have been performed with similar results.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Table 1.
Source data
Source Data Fig. 1
Unprocessed western blots and gels, and statistical source data.
Source Data Fig. 2
Unprocessed western blots and gels, and statistical source data.
Source Data Fig. 3
Unprocessed western blots and gels, and statistical source data.
Source Data Fig. 4
Unprocessed western blots and gels, and statistical source data.
Source Data Fig. 5
Unprocessed western blots and gels, and statistical source data.
Source Data Fig. 6
Unprocessed western blots and gels, and statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and gels, and statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and gels.
Source Data Extended Data Fig. 3
Unprocessed western blots and gels, and statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Unprocessed western blots and gels.
Rights and permissions
About this article
Cite this article
Ji, S., Siegel, A., Shan, So. et al. Chloroplast SRP43 autonomously protects chlorophyll biosynthesis proteins against heat shock. Nat. Plants 7, 1420–1432 (2021). https://doi.org/10.1038/s41477-021-00994-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-021-00994-y